

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
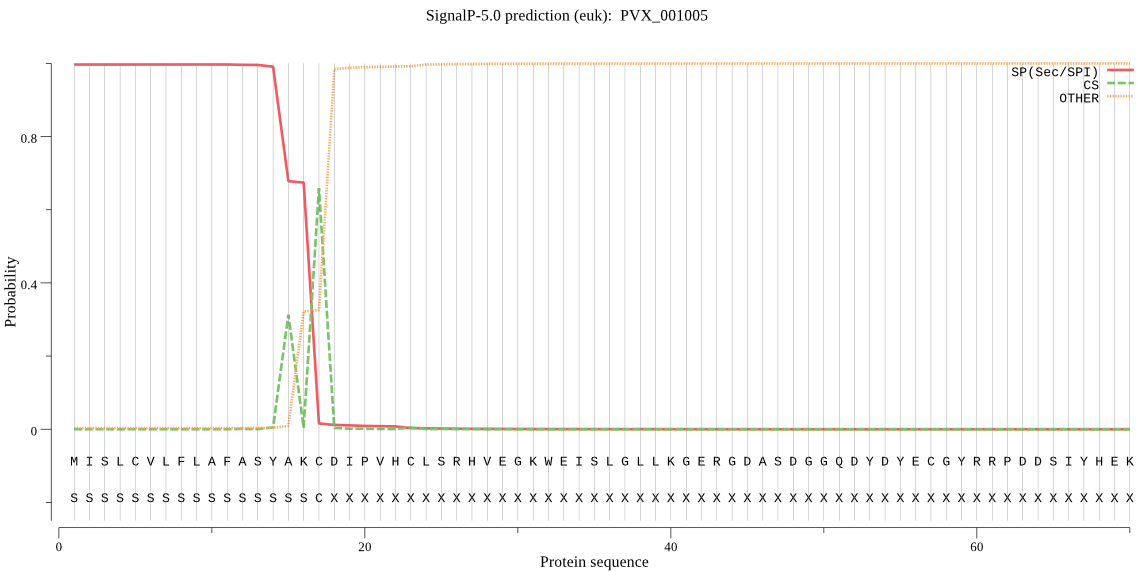
| PVX_001005 | SP | 0.001622 | 0.998362 | 0.000017 | CS pos: 17-18. AKC-DI. Pr: 0.9138 |
MISLCVLFLAFASYAKCDIPVHCLSRHVEGKWEISLGLLKGERGDASDGGQDYDYECGYR RPDDSIYHEKLDPDHLKNRFEEKKKQIILFNTDRTIQIIENGNVNPQYSGFWRIVYDEGL YMEVYKQKEEKEIYFSFFKFERRNDASYSYCNRLIMGVVNVYQLSGSGGGSSSGRPRVDG LQDISEGREKGPQMWDSFLHNMGHKSTNGVPPNRSSLSKESFPSEGSKGVRKSPTHSIDY YNDNYTQFDLFTVKRFCWYGKKVGTPDEPTNKIPVEIISPLVLDPDTHNKNYANVKLEWG GGTAEGTQRGGHLEVHTNQTATQWRAPPTHTSKSHKGVKPNSIFNTYREKEVPLAQFDWT NEADVRKRLNGKWVKVIDEAIDQKECGSCYANSAALIINSRVRIKYNYIKNIDRISFSNK QLVLCDFFNQGCNGGYIFLSLKYAYENFLFTNKCFVRYFNLYLNRDKKNSALCDRFDTFK TFLQKGRGGDASSGPMGPARIQIKQVRQHGHFGGGEEKVTGEELVREEDLDGGEIGKSST HDEQPKGGISHNGENKLTGRHVRDVRGGRDLPGEDQLNEGYLPVDPAAKDASEVDVLKLH ECDAKVKVTKFEYLDIEDEEDLKKYLYYNGPVAAAIEPSKNFSAYREGILTGKFIKMSDG GESNAYVWNKVDHAVVIVGWGEDTVENLKKKKKKIHHYSGHLDAYAGGKGGEAAGKVVKY WKILNSWGTHWGYDGYFYILRDENYFSIKSYLLACDVSLFLR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_001005 | 539 S | IGKSSTHDE | 0.99 | unsp | PVX_001005 | 539 S | IGKSSTHDE | 0.99 | unsp | PVX_001005 | 539 S | IGKSSTHDE | 0.99 | unsp | PVX_001005 | 233 S | GVRKSPTHS | 0.996 | unsp | PVX_001005 | 334 S | HTSKSHKGV | 0.995 | unsp |