

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_001780 | SP | 0.012362 | 0.987438 | 0.000200 | CS pos: 30-31. GEG-KW. Pr: 0.6636 |
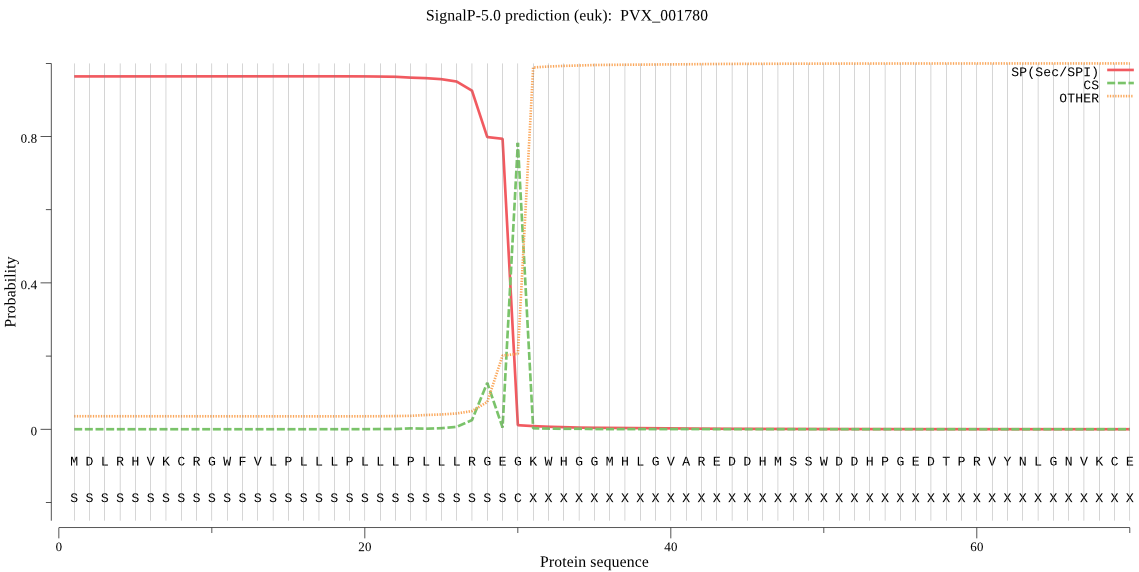
MDLRHVKCRGWFVLPLLLPLLLPLLLRGEGKWHGGMHLGVAREDDHMSSWDDHPGEDTPR VYNLGNVKCEQLEEETYGGGLLGSPKGETNPSGMNTVQWITWKGKRDGGEAAPLGGGPPY SCVDMLPSRHKVTKRMLSSVMGDHLEVPTSGGHSPDGSFQGSVLNVHFTENKNLYTNVKV GGQPLKLALNSRVEGIYVFMKDSQACHVDEEGEEKNRVCYDPNTSRNSTWCNNDLLCLPA ILSKSYECYSDSNLLLENKAEYPSMYYDSLKFTESHVEGSDDVEVLDLVKAAGAEKAAGE GDAAGGGDAAGGGDAAGGGEEVGYANAAGGETDATFQNTDVKLVTDLSLYKGWSLFKDTD GMMGLAGRELSCRNVSAWNTIVEKSNSLYALDVNLPEGSVKPFVESPTQEDSKRGASSYA KFVPKKASSNEADGAAEPSGSTSEIHIGDYKKSFGPIVWSEPRERGGIFSDSFMQFTLYN LEVCQNNIFGKYSSNWQGVIDLSSKCLVLPKMFWLSLMEYLPVNKNDERCIPKRKEVTFD EGTVPRMCAVDPGSRPLPVLKFYLSDNDTVSDNNIGGGSGGEIGGDPPRGGIQEVHIPLD NLIINEEGQNDGYLCVLPDVHEGVSSENSGRTTKPLIKFGTYVINNFYVVVDQENYRVGF ANKKEYHSSNDRCTQRAQCVGNQFYEPALNICVDPDCSVWYFYTLNEETKKCESISSRFY VFLLIILLLLLLDIQSYYFYRKSVHVAKVSSR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_001780 | 138 S | KRMLSSVMG | 0.994 | unsp | PVX_001780 | 138 S | KRMLSSVMG | 0.994 | unsp | PVX_001780 | 138 S | KRMLSSVMG | 0.994 | unsp | PVX_001780 | 154 S | SGGHSPDGS | 0.997 | unsp | PVX_001780 | 324 Y | EEVGYANAA | 0.99 | unsp | PVX_001780 | 668 S | KEYHSSNDR | 0.994 | unsp | PVX_001780 | 49 S | DHMSSWDDH | 0.996 | unsp | PVX_001780 | 58 T | PGEDTPRVY | 0.996 | unsp |