

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_003790 | SP | 0.295496 | 0.701652 | 0.002852 | CS pos: 24-25. VKT-NI. Pr: 0.9313 |
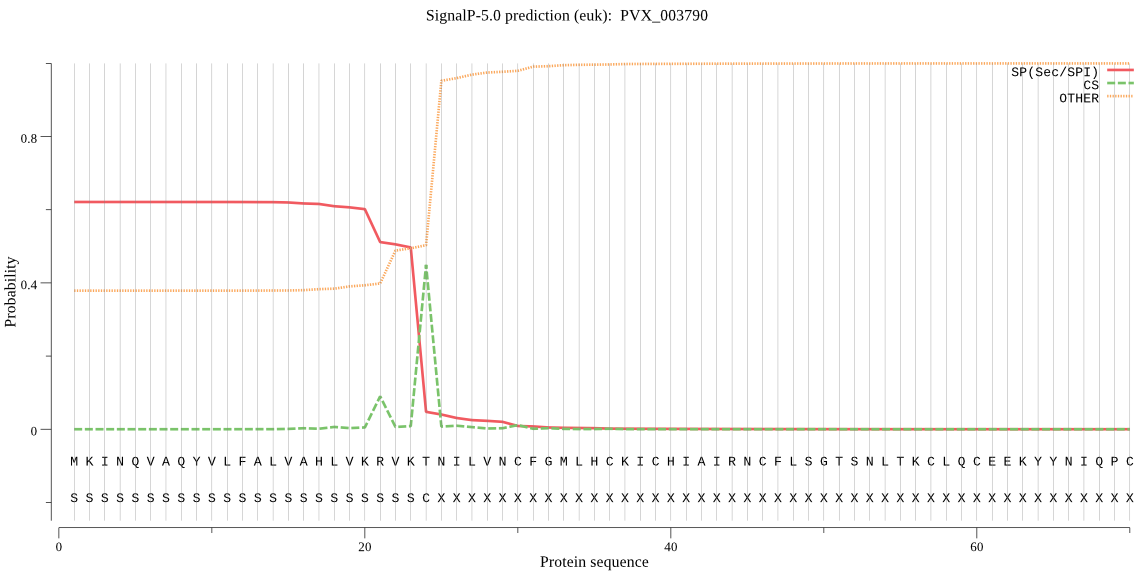
MKINQVAQYVLFALVAHLVKRVKTNILVNCFGMLHCKICHIAIRNCFLSGTSNLTKCLQC EEKYYNIQPCTHQTENFLKTSRDKGAFVEMKDHDYLTDGKVDDLISEVVKISLERQKNAV PAAEDRNEDFHKKIMQLCLYSNFNDNYENAKKHTQASVEEVERHIHKIINMYMRESNNME HIINSLKNPALCLKNPAEWTKDRAGYKDNDVPSVGIIPERKLFKPYEVKSLMSSLYSYKS NCNRQFCDRFSDPNECEHSIRVLNQGTCGNCWAFASSQVISAYRCRKGLGFAEPSVKYVT LCRNRHSDDYEENPLGHYNDNICREGGHLSYYLDTVDKTGMLPTSYDVPYNEPLRGADCP ESTAKWANIWEKVNPLTRILNGYLYRGYFKISFHEYARSGKTQELISLIKDYIMDQGSLF VSMEVNEKLSFDHDGEKVMMNCEYKASPDHALTLIGYGDYIKPSGEKSSYWLIRNSWGSH WGDKGNFKVDMYGPPNCNGAVLCNAFPLLLQMNGKKITKPLPKDLASTDNKVRYDHSIFN RDNNRAQSRRYNQNREQDRPNGRADNNPFDNPKYNEIYDDDDRDAPAFDPFRGDYVYPHM DNANGAERGNEGMNDNRGGKIRRKVFKTSLVVTIGDTQFKRDIFMKRKEEYKEQHSCLRT YSLDSLADVSCRNNCEQYIDLCKHYTSIGRCLMRFANNYKCVYCGM
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003790 | 157 S | HTQASVEEV | 0.996 | unsp | PVX_003790 | 157 S | HTQASVEEV | 0.996 | unsp | PVX_003790 | 157 S | HTQASVEEV | 0.996 | unsp | PVX_003790 | 237 S | SSLYSYKSN | 0.992 | unsp | PVX_003790 | 64 Y | CEEKYYNIQ | 0.99 | unsp | PVX_003790 | 112 S | VVKISLERQ | 0.991 | unsp |