

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
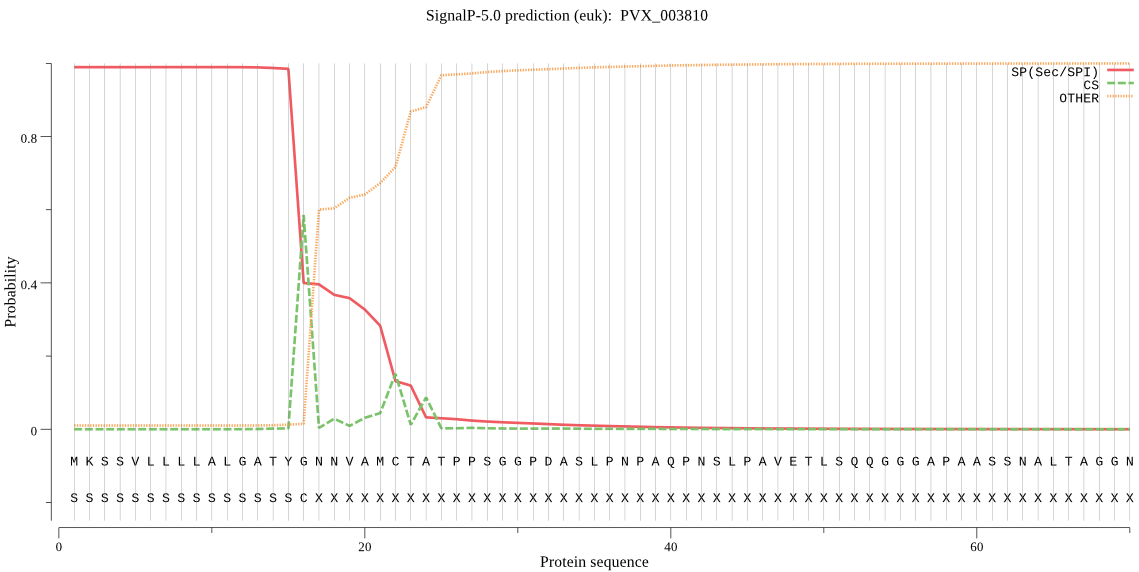
| PVX_003810 | SP | 0.000192 | 0.999806 | 0.000003 | CS pos: 16-17. TYG-NN. Pr: 0.7545 |
MKSSVLLLLALGATYGNNVAMCTATPPSGGPDASLPNPAQPNSLPAVETLSQQGGGAPAA SSNALTAGGNVPPGSQVNSGEQGGATQLQATPKKAELQSSLLKNFTGVKVTGPCDTEVGL YLIPYIYISVNAKTDTIELSTRFPNSDNVLVQFKKEGETLTNVCEGSSEKTFKFIMHLED SILTLKWIVNPQSTTANKNKADVKKYKVPELERPITSIQVHSVLIQEDTVIYKSKDYSVK TDIPEKCEQVASACFLSGNTDIESCYTCNLLIHNEDTSDKCFDYVSSEFKNQFQDIKVKG QDDEESSEYKLAQAIDAVLSGIYKKDQNGKKELLTWGEVDPNVKEQISSYCHLLKDVDAS GSLEVHQMGSEMDIFNNLITLLQKHSEEQKATLEWKLQNPALCLKDANNWVVNKKGLLLP LLQNGGSAINFGESNHVEDVKKDNLSTYQEGPDGVIDLSVVHQNAHASSTPFTNHMFCNS DYCDRAKDTSSCMAKIEVQDQGDCSTSWLFASKVHLEAIKCMKGHDHVGASALYVANCSN KEAKDKCHSPSNPLEFLNTLEETKFLPAESDLPYSYKAVNNVCPEPKSHWKNLWENVKLL GPTNEPNSVSVKGYTAYLSDHFKGNMDAFIKSVKSQVMSKGSVIAYVKADDQMSYDLNGK KILSLCGSETPDLAVNIVGYGNYITAEGQKKSYWLLQNSWGKHWGDKGTFKVDMHGSAHC QHNFIHTAVVFNLDVPLSQSPAKDTELYSYHLKSSPDFYKNLYYNAVGGEKGSVLSNAVQ GQDTPQEEGTSPGTAAEGEGRPGPVGPGGQQPQEHASSAGGPNSNEQQLDVARQQQQEGA EKESSQQLPQQQPLTQSQQSTQPGVGGAASTPLSQGTPGQANSNGEAENANISQIIHVLK HIKKTKMVTRIVTYEGEYDLGDHSCSRTQASSLEKLDDCIKFCNDNLSMCERTVSTGYCL TKLRKANDCIFCFV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003810 | 635 S | KSVKSQVMS | 0.991 | unsp | PVX_003810 | 635 S | KSVKSQVMS | 0.991 | unsp | PVX_003810 | 635 S | KSVKSQVMS | 0.991 | unsp | PVX_003810 | 932 S | TQASSLEKL | 0.993 | unsp | PVX_003810 | 238 S | SKDYSVKTD | 0.992 | unsp | PVX_003810 | 549 S | DKCHSPSNP | 0.994 | unsp |