

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
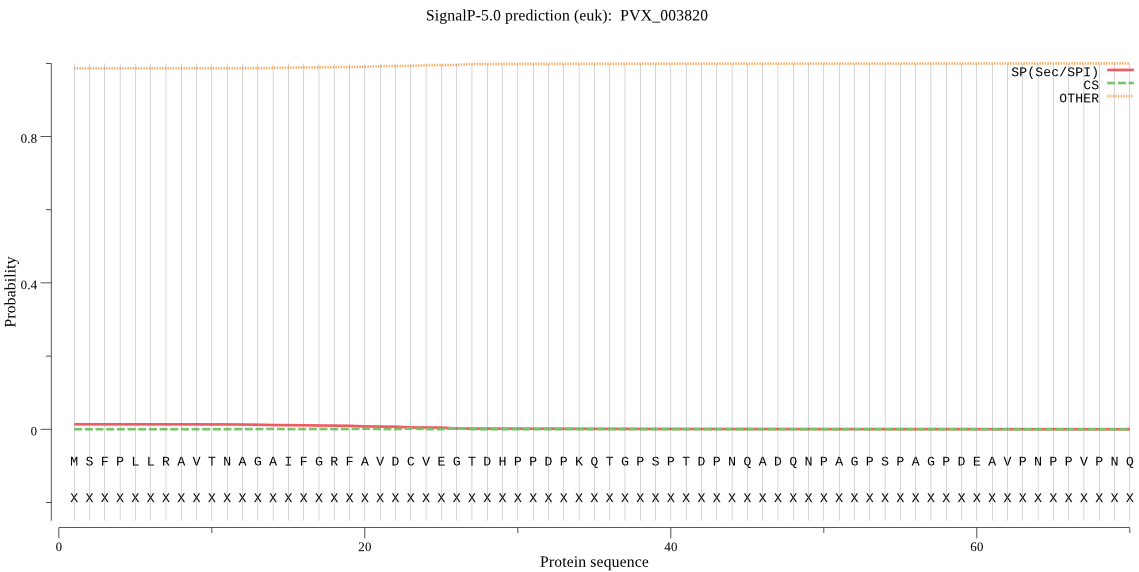
| PVX_003820 | OTHER | 0.910741 | 0.043359 | 0.045900 |
MSFPLLRAVTNAGAIFGRFAVDCVEGTDHPPDPKQTGPSPTDPNQADQNPAGPSPAGPDE AVPNPPVPNQPVRVPPPPTPPSEHNVSPAGQTPGGTPQVETEADQINLPLSNDVPGSEMS VISLPDDDNEEEAKGPSGTHEPTQGADKPLEISITSALLKDHNGVKITGPCSADFQLFLV PHIIIHVESAKDKIELRSKWEDVDNKQSKGAKVPVEELKSGILFKQPNETLQNNCAEGKN FKFVLYVHEDQLIVKWKVYNKTPAATDNKKVDVRKYLVKNLKRPITTIHVHSSSVNQTTF LLESKSYALKKDIPDQCDAMVTNCFLSGILDVEKCYHCSLLVSSKDTSNMCFNYASPDIK EKFNEIKVEGQDDEESSEYKLAQAIDAVLSGIYKKDQNGKKELLTWGEVDPNVKEQISSY CHLLKDVDTSGTLDVHQMGSQEHIFNNLITLLQKHAEEKQESLQNKLKNAAICLKQAHNW VANKVGLALPQLAYNPGKGVEQVEENPPNGESANGESAHDHTHEQGFDGVIDLPPFGTSG AQASVYSDSMYCNGDYCDRAKDSSSCMAKIEAGDQGDCANSWLFASKAHLEAIQCMKGHD HVGASALYVANCSGKEAKDKCHVASNPLEFLNTLEETKFLPAESDLPYSYKAVNNVCPQP KSHWKNLWANVKLLEKQYEPNAVSTKGYTAYQSDHFKGNMDAFIKTVKSELMKKGSAIAY VKAAGALSYDLNGKKVLSLCGSETPDLAVNIVGYGNYISAEGQKKSYWLLQNSWGKHWGD KGTFKVDMLGPDHCHHNFIHTAAVFNLDMPQANPPPVEFQLSNYYLKSSPDFYTNLYYKN VKATPGGAKVYGSEGESEVTEGAQNRGAQTAQDRGKVSPGTEQPEKAGGNAAPEAAQITE GGGNRASEASPGIAVPPTPQDPAGGDVSAGTGVESPVSLPQAAGGTPSASLNTQESSSQA QDPQSPAREEQVAAPPQPDVPTPTPQQASTSALVKAPLAQSADAESKEVLHILKHVKDNK VKTNLVTYSTTKSITDDHSCSRSHAQDPEKQSECIDFCEKNWNACADKISPGYCLMQKRG KNDCIFCYV
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003820 | T | 79 | 0.665 | 0.629 | PVX_003820 | T | 41 | 0.626 | 0.469 | PVX_003820 | T | 92 | 0.574 | 0.477 | PVX_003820 | S | 82 | 0.563 | 0.270 | PVX_003820 | S | 87 | 0.556 | 0.106 | PVX_003820 | T | 36 | 0.528 | 0.464 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003820 | T | 79 | 0.665 | 0.629 | PVX_003820 | T | 41 | 0.626 | 0.469 | PVX_003820 | T | 92 | 0.574 | 0.477 | PVX_003820 | S | 82 | 0.563 | 0.270 | PVX_003820 | S | 87 | 0.556 | 0.106 | PVX_003820 | T | 36 | 0.528 | 0.464 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003820 | 343 S | SLLVSSKDT | 0.995 | unsp | PVX_003820 | 343 S | SLLVSSKDT | 0.995 | unsp | PVX_003820 | 343 S | SLLVSSKDT | 0.995 | unsp | PVX_003820 | 853 S | KVYGSEGES | 0.994 | unsp | PVX_003820 | 965 S | QDPQSPARE | 0.992 | unsp | PVX_003820 | 39 S | QTGPSPTDP | 0.996 | unsp | PVX_003820 | 189 S | IHVESAKDK | 0.996 | unsp |