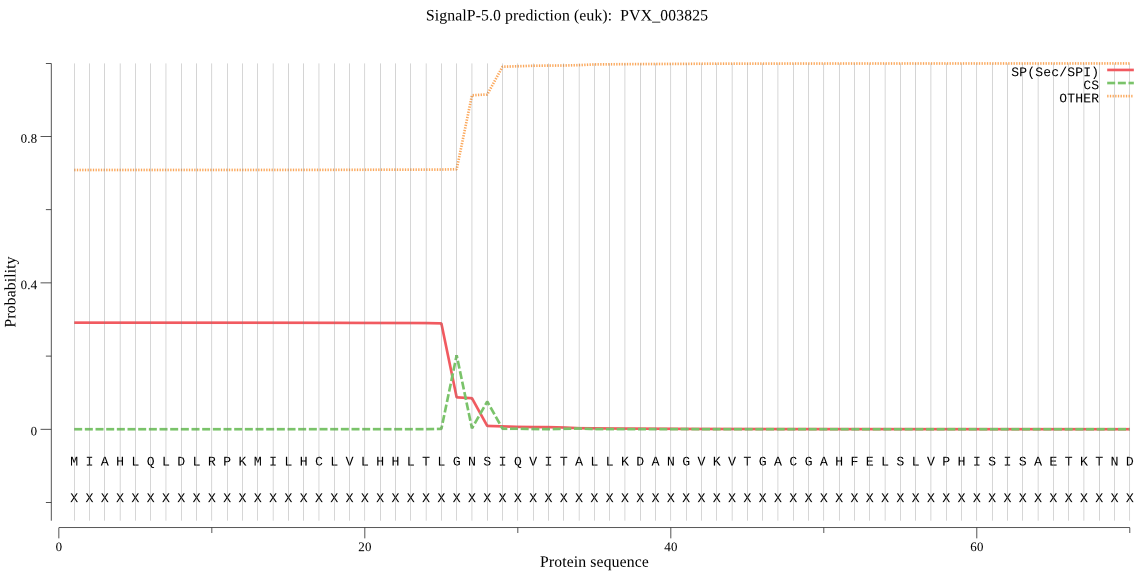
Fasta :-
MIAHLQLDLRPKMILHCLVLHHLTLGNSIQVITALLKDANGVKVTGACGAHFELSLVPHI
SISAETKTNDIKLRPILHKLLDAKEADVGTTKLDNAIQFEPNQEKLKNKCQPGKENEDTF
KFLVFIHEGELTLKWKVYNKSAPADASKEVDVRKYTLKNLDQPITTVQVRSSKVDEETLL
VESKSYYVNRDIPDKCDVIATDCYLNGNVDIEKCFQCTLLMENSDTTNECFKYVSSQVTD
KFKDIKVNAQDEEDPAEVELAASIGKILQGVFKKGEAAHIELLTFDEVDAALKEELLNYC
SSMKEVDASGVLDQYQLGSEEDIFANLTSILKNHAGETKSTLQNKLKNPAICLKNADEWV
ESKKGLLLPSLSHTNGEVTPPETPKEDHMSDADEGQQSVPYDAVINLVSAEEKNNQSSLI
EDSTFCTEDYCNRWKDDTSCVSKIEAEDQGVCSTSWLFASKVHLETIKCMKGYDHIATSA
LYVANCSSKEAKDKCHAASNPLEFLDILEETQFLPAESNLPYSYKAVSNACPEPKSHWQN
LWENVKLLEKQYEPNAVSTKGYTAYQSDHFKGNMDAFINLVKSQVMSKGSVIAYVKADEL
MGYDFNGKNVHSLCGSETPNHAVNIIGYGNYVSAEGVKKSYWLLRNSWGKYWGDDGNFKI
DMHGADHCQHNFIHTAAVFNLEMPLVQSAAKKDNHLYSYYLKGSPDLYQNMYYKGFGSQG
GNAKGGEKGQVGTPVVSGQEEEEVASDRQDQVREQSHVEASAGPDGLQAGQVDVKAEEED
TKAGQGGEDSAATQDGGSHGPGAVQPLAEQSDTRGVASNLPDGSQLPGSGHEAGQSGHGV
AESGSEVPESRPEAPPGEPGAAHSEPQVTESEHGVPPSGSGVPPGGPGVPPSGPEVGRSE
TGETLSQPAESPPLPNPPSNTAKITQVLHILKHVKQGKVKTRLVTYDSNAAMSADHVCSR
AVASDPEKQDECVKFCDDHWNDCKDKISPAYCLTQIRGKKDCFFCYI