

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
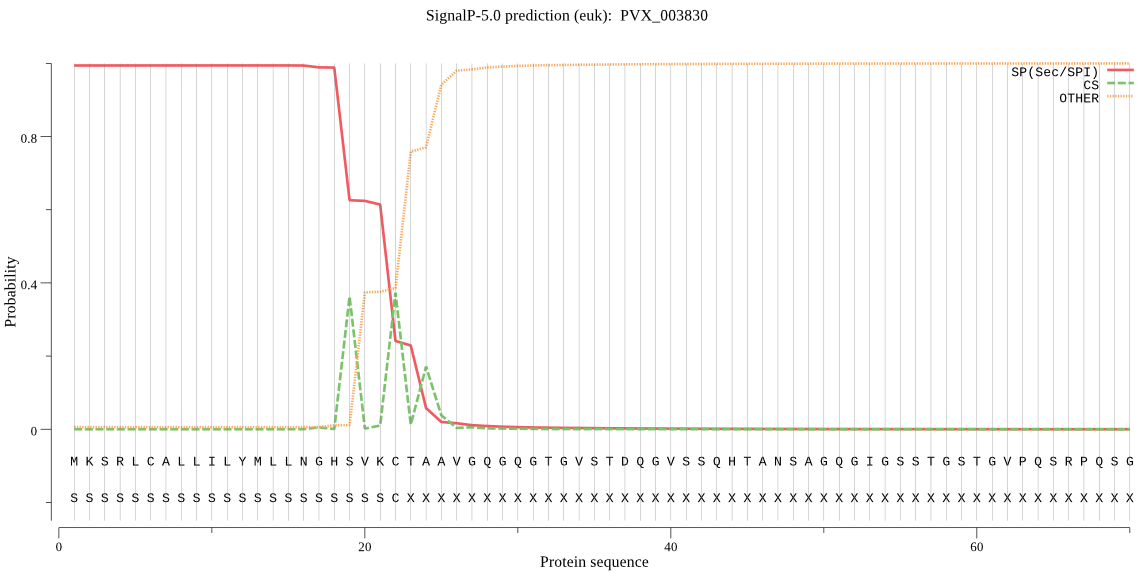
| PVX_003830 | SP | 0.003677 | 0.996304 | 0.000019 | CS pos: 22-23. VKC-TA. Pr: 0.5187 |
MKSRLCALLILYMLLNGHSVKCTAAVGQGQGTGVSTDQGVSSQHTANSAGQGIGSSTGST GVPQSRPQSGGGDTGTQGGQQDRASGTSATHVPVVPQNGHVQSNPPPSTSSGGPNGGGGA SITQNVQQANLQAGSDTQVAATPSESFTNPIQVKASLLRDQKGLKITGPCKSYFQVYLVP YLYLNVNAKESEIEMDPMFMKVDDKIKFEKEKHLLNNICEDNKTFKLVVYMYEGELTIKW KVYPPKGETSSDKTLDIRKYKMKDIGQPITSMQVVVMSELNKTIYMESKNFAVMNQIPEK CDAIANECFLSGVLDVQKCYHCTLLLQKKENAEECFKFVSPTIKNRFEDIQTKGEDEENP NVVELEETIDLLLNKIYKNGEGESNEVDQLAIIDSSFQSDLLKYCSLMKEVDTSGSLDNH QLGDAEDVFANLTHLLQSNSDHDVPSLKNKLKSPAICLKNVGHWVGSKTGLVLPTLEYST SQDNAESFDEGEEDTSNSTTTQSADVHPLNVSDKLFCNDEYCDRAKDSSSCIAKIEAGDQ GDCSTSWLFASKVHLEAIKCMKGHDHVASSALYVANCSGKEANDKCHAASNPLEFLNTLE ETKFLAAESDLPYSYKAVNNACPEPKSHWKNLWENVKLLDPTNEPNSVSTKGYTAYQSDH FKGNMDAFIKLVKSEVMKKGSAIAYVKAQGALSYDLNGKKVQSLCGGETPDLAVNIVGYG NYITAEGQKKSYWLLQNSWGKHWGDDGNFKVDMHGPDHCQNNFIHTAAVFNLDVPVVAPA PSSDPEINDYYMKNSPDFFSNFYFNKYEAEKANGLGEEKGPVNNSVLYGQSGEETSALPA SVGDQSGKVVAQTAEGLGAQQGSGGGLGAAAGPTGQEGVGVAAPGGRGTGGEAIAAAAVV VVGGGGSGSAGERGTGVGVAPGVGAAGRSVGEGQQLPAGAASPGTGTQHVGGSVSGGTNS GNGATGNSGPNTQRGQVSQTVNEAAPSTVEKPKSIDSTGVISEGITGVFHFLKNVKKGKV SSNFVTYDNTKAIGDKACSRVQSSDVDKLDDCVKFCEANWNECKGKVSPGYCLTKKKGNN DCFFCFV
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| PVX_003830 | T | 109 | 0.547 | 0.451 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| PVX_003830 | T | 109 | 0.547 | 0.451 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003830 | 438 S | HLLQSNSDH | 0.991 | unsp | PVX_003830 | 841 S | ALPASVGDQ | 0.995 | unsp |