

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
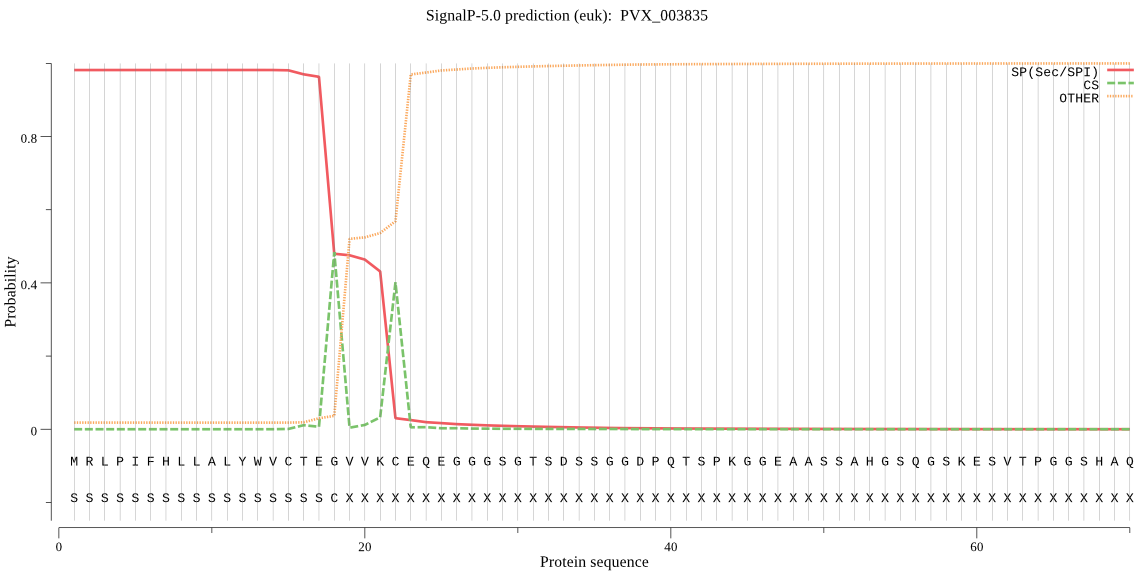
| PVX_003835 | SP | 0.014595 | 0.985403 | 0.000001 | CS pos: 18-19. TEG-VV. Pr: 0.4925 |
MRLPIFHLLALYWVCTEGVVKCEQEGGGSGTSDSSGGDPQTSPKGGEAASSAHGSQGSKE SVTPGGSHAQQEAAAPEKTLVPQESPAKEGGAPPSVPENQAHSPQTPGGPPPTASNIIGT GVSNNGSPPHAAATPGGDNPKQEEATTALKPIQVKSALLKDHKGVKVTGPCDATFQVYLV PYLYIDVNAKNTEIEMDPMFTKVDNKIKFEEGKHRLMNICEKGKTFKLVVYMYEDVLTIK WKVYAPKGVPAADKTLDVRKYKMRDIGRPITSIQVLATSKNDETVLVESKNYSVMEEIPE KCDAIANECFLSGVLDVQKCYHCTLLLEKKENAQECFKFISPEIRDRFDDIKTKGEDEED PNEAELEESIHTILEKMYQNGGANNEVNHLAILDDTLKEEIQNYCAMMKEMDTTGVLENY EMGNADDVFGNIAKMMGKNGNYSVSSLPNKMKNAAICLKSPDDWVEGKTGLMLPNLVPPY LVHNNVEDVTTSLETSSDGTTAFKQDDDGTIDLTNLDCVDVHPSVLADKMHCNDDYCDRA KDTSSCMAKIEVEDQGDCATSWLFASKMHLETIKCMKGYDHVASSALYVANCSDKEESDK CQAASNPLEFLDILEETKFLPAESDLPYSYKQMGNACPEPKSYWQNLWADVKLLEKQYEP NAVSTKGYTAYQSDHFMANMDAFIKMVKSEVMKKGSAIAYVKAAGVMTHDMNGVDVHSLC GGETPDLAVNIVGYGNYISPEGVKKSYWLMRNSWGKYWGDDGTFKVDMHGPPGCQHNFIH TAAVFNLDMPLATHSINKDPEISDYYLKNSSDYYENLYYKKFQGSGATGGAGKQWVQGAS TVYGQEGDPKVADEAGGEAAQSITTHSRAEPAGDAPPSGPPHAGTDSEQGEGTGPTDGEG PTRVVDEKAQAGGPLTSPGTLQDAVAPGGPQGPPEQGSPGSPGPQETPGPQGPPVQQGSP GPQLPPVQPTAPVPPVLPVPPVLPVPPAAPLPGAPTNSTVPTTESEVKEVLHILKHIKKG KVKVGLVTYDTEKVIGAEKVCSRAYAVDHEKQEECVEFCEANWDACKGKVSPGYCLTKKR GSNDCFFCFV
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003835 | T | 106 | 0.629 | 0.277 | PVX_003835 | T | 120 | 0.599 | 0.215 | PVX_003835 | T | 113 | 0.541 | 0.425 | PVX_003835 | T | 41 | 0.521 | 0.178 | PVX_003835 | T | 79 | 0.503 | 0.079 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003835 | T | 106 | 0.629 | 0.277 | PVX_003835 | T | 120 | 0.599 | 0.215 | PVX_003835 | T | 113 | 0.541 | 0.425 | PVX_003835 | T | 41 | 0.521 | 0.178 | PVX_003835 | T | 79 | 0.503 | 0.079 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003835 | 460 S | ICLKSPDDW | 0.991 | unsp | PVX_003835 | 460 S | ICLKSPDDW | 0.991 | unsp | PVX_003835 | 460 S | ICLKSPDDW | 0.991 | unsp | PVX_003835 | 598 S | DKEESDKCQ | 0.995 | unsp | PVX_003835 | 1082 S | KKRGSNDCF | 0.993 | unsp | PVX_003835 | 35 S | TSDSSGGDP | 0.996 | unsp | PVX_003835 | 42 S | DPQTSPKGG | 0.998 | unsp |