

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
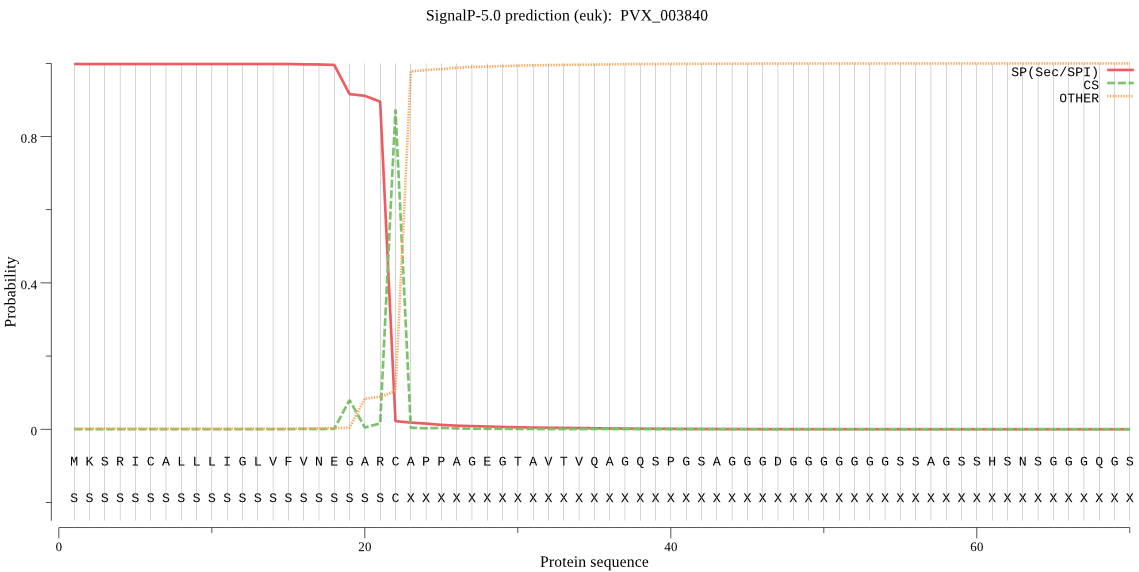
| PVX_003840 | SP | 0.001279 | 0.998721 | 0.000001 | CS pos: 22-23. ARC-AP. Pr: 0.8837 |
MKSRICALLLIGLVFVNEGARCAPPAGEGTAVTVQAGQSPGSAGGGDGGGGGGGSSAGSS HSNSGGGQGSDAAAQRPAAPQPPPTPPAEGSSSPPAGNQNPSTSGSQPQNVSSVAAPVPP VEATPPPSVTNPFKVKSSLLKDQKGLKITGPCESYFQVYLVPYLYMNVNATSSEIEMEPM FMKVDDKIKFEKEKHLLHNICAADKTFKLVLYMYDGVLTIKWKVYPLNGDKTADRTLDIR KYKMKDIGQPITSMQVVVMTEQNKTVYVESKNFSIMNEIPEKCDAIANECFMSGVLDVQK CYHCSLLLQKKESAQECFKFVSPKIKNSFDEIQAKGEDEENPNEEELQKTIDNIFVKVYK KGENLYKEVDQLAIIDSSFKSELLKYCSLMKEVDTSGALDNHQLGNAEDVFAHITTMLHS NSDLDVFSLKSKLKNAALCLKNGDEWVGSKTGLVLPNLSPQNFEDSPTQTFDGGDEQAAL EAGEDGVVDLTSLQSVDVSPFLVTDKLFCNDDYCDRAKDTSSCVAKIEVQDQGDCATSWL FASKVHLETIKCVKGYDHVGASALYVANCSGTEANDKCHAASNPLEFLNTLEETKFLAAE SDLPYSYKAVNNACPEPKSHWKNLWENVKLLDPTNEPNSVSTKGYTAYQSDHFKGNMDAF IKLVKSEVMKKGSAIAYVKAQGALSYDLNGKKVLSLCGGETPDLAVNIVGYGNYISAEGV KKPYWLLQNSWGKHWGDKGTFKVDMHGPPGCQHNFIHTAAVFNLDIPVVVPAPNSDPEIN NYYLKNSPDFFSNFYLNKYEAGSDGNSDDAKSVSGNSTVQGPDGPAGPSGPDGNVGQKGA AARTGEPSTAVERSQTADGVTTPPGGGGANGGRQGAEASVQERNAVDSGSPQAVTRPTPP PERVASGLASVQHSGDTTLPQSTTSSVTQNPPVATRPAPQTTHQTAPAQPATPREPLSSL KGSTGVNVTEVKEALHFLKSVKNGKVKSNFVAYVNADAMGDEKVCSRAFSTDADKQTECI EFCEKNWDACKGKVSPGYCLTKKRGSNDCFFCFV
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003840 | T | 85 | 0.665 | 0.534 | PVX_003840 | S | 92 | 0.625 | 0.286 | PVX_003840 | S | 91 | 0.623 | 0.307 | PVX_003840 | S | 93 | 0.621 | 0.100 | PVX_003840 | T | 103 | 0.603 | 0.098 | PVX_003840 | T | 124 | 0.574 | 0.672 | PVX_003840 | S | 106 | 0.519 | 0.032 | PVX_003840 | S | 113 | 0.509 | 0.051 | PVX_003840 | S | 102 | 0.504 | 0.110 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003840 | T | 85 | 0.665 | 0.534 | PVX_003840 | S | 92 | 0.625 | 0.286 | PVX_003840 | S | 91 | 0.623 | 0.307 | PVX_003840 | S | 93 | 0.621 | 0.100 | PVX_003840 | T | 103 | 0.603 | 0.098 | PVX_003840 | T | 124 | 0.574 | 0.672 | PVX_003840 | S | 106 | 0.519 | 0.032 | PVX_003840 | S | 113 | 0.509 | 0.051 | PVX_003840 | S | 102 | 0.504 | 0.110 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003840 | 420 S | TMLHSNSDL | 0.994 | unsp | PVX_003840 | 1046 S | KKRGSNDCF | 0.993 | unsp |