

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
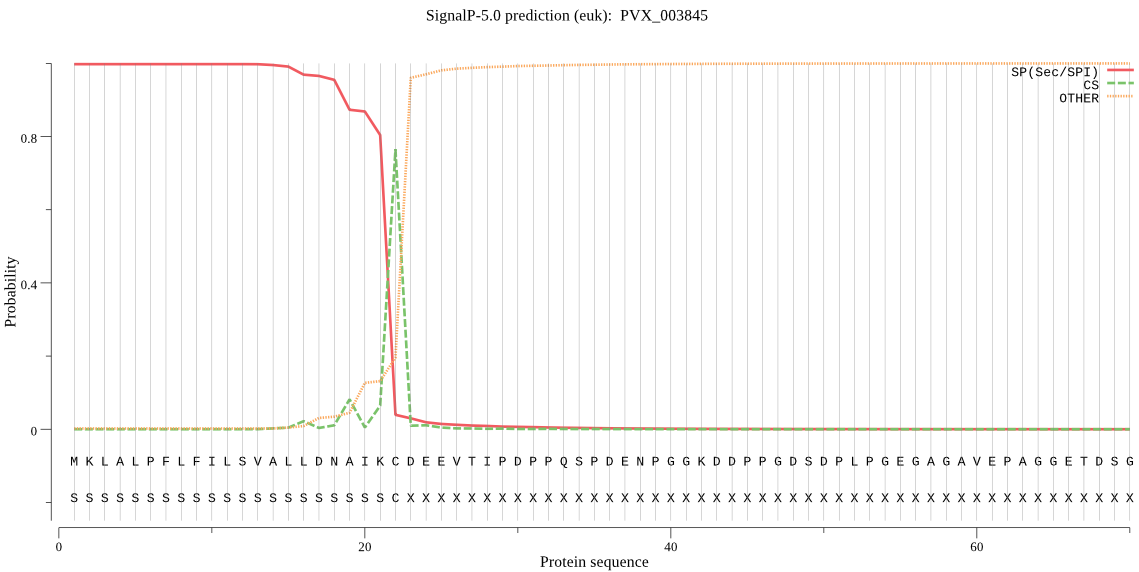
| PVX_003845 | SP | 0.000567 | 0.999433 | 0.000000 | CS pos: 22-23. IKC-DE. Pr: 0.8244 |
MKLALPFLFILSVALLDNAIKCDEEVTIPDPPQSPDENPGGKDDPPGDSDPLPGEGAGAV EPAGGETDSGVEEGPAEQAVDQPLTQSTDQPADQPAEQPADQALTQPTDQPANQPVDQPT DQPIDQPTDQPVDQTTDQTTEQPAGEPLTQSTDEPVDQPLTQSTDQPADQSADQPVDQTT DQVTEQPTDQPTEQPTDQTTDQPTEQPTDEPLTQPTDEPLPQPIVEASDRAAAAAVKNPN EIEAKCAQLKDQDGVKITGPCGAKFQVFLIPHVTINVETETNAIHLGKKLDDVVITKKMH KGVGGKSPPLLQFEEDADSLLNQCTEGKTFKFVVVVKGEELILKWKVYEKVPSPSDNNKV DVRTFLLKNTDRPITAIQVHTAKGNEDSFLLESKEYILADDMPAQCDLIAANCFLSGSLD IEGCYKCALLSENAELSSPCFDYLSPDVKNDYEEIKRKAQQQGDLKEVQLAASIGKILQG VFKKGEAGLNELVTFDQADAALKEELLNYCALMKEVDASGVLDQYQLGSEEDIFANLTSI LKNHAGETKSTLQNKLKNPAICLKNANEWMESKKGLLLPSLSYTNVEATLPENAPEKEDP PKGSQKIQTNGYDGIINFDSNEETNMQSTSFIDNMYCNDEYCDRWKDSSSCVAKIEVEDQ GACSNSWLFASKVHLESMKCMNGHDHMATSALYVANCSGKEEKDKCHVASNPLEFLDILE ETQFLPAESDLPYSYKAVNNVCPQPKSHWQNIWADVKLLDKQDDPNAVSAKGYAAYQSDH FKGNMDAFIKLVKSEVMKKGSVIAYVKADDQMSYDLNGKKVLSLCGSEEPNLAVNIVGYG NYISAEGVKKPYWLLRNSWGKHWGDDGTFKVDMHGPPGCQHNFIHTAAVFNLHIPPMENV EKKKPLLYNYYMKSSPDFYNHIFYRGVQTGEESEMGISGQEKVSISAVSANTSAADTLDG VDHSSVVVEGKEKNPAGEGESAQGVESPPEAEEQDEEEGDAESEEEGEDEPEEEGDGEQE EEGDDESEEVDEEAEEAGAEAEEGEEESEEGGVEAEEGDSEAANNDVDAAEPSQVAAPGA SPGGEKPQTVAPPSASNPATPPSAAPAPTRNASLIKVKQITEVIHIMKHIKSGKLRFGIA TYEDDLGIANNHDCLRSYSQDPEKLPECIQFCYDEWNNCKGAPSPGYCLNQRRRKNDCYF CFV
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003845 | T | 200 | 0.588 | 0.248 | PVX_003845 | T | 199 | 0.584 | 0.026 | PVX_003845 | T | 204 | 0.580 | 0.555 | PVX_003845 | T | 208 | 0.576 | 0.541 | PVX_003845 | T | 196 | 0.571 | 0.421 | PVX_003845 | T | 213 | 0.561 | 0.260 | PVX_003845 | T | 192 | 0.534 | 0.555 | PVX_003845 | T | 216 | 0.501 | 0.455 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003845 | T | 200 | 0.588 | 0.248 | PVX_003845 | T | 199 | 0.584 | 0.026 | PVX_003845 | T | 204 | 0.580 | 0.555 | PVX_003845 | T | 208 | 0.576 | 0.541 | PVX_003845 | T | 196 | 0.571 | 0.421 | PVX_003845 | T | 213 | 0.561 | 0.260 | PVX_003845 | T | 192 | 0.534 | 0.555 | PVX_003845 | T | 216 | 0.501 | 0.455 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003845 | 529 S | YQLGSEEDI | 0.99 | unsp | PVX_003845 | 529 S | YQLGSEEDI | 0.99 | unsp | PVX_003845 | 529 S | YQLGSEEDI | 0.99 | unsp | PVX_003845 | 987 S | QGVESPPEA | 0.996 | unsp | PVX_003845 | 1003 S | GDAESEEEG | 0.997 | unsp | PVX_003845 | 34 S | DPPQSPDEN | 0.996 | unsp | PVX_003845 | 353 S | EKVPSPSDN | 0.996 | unsp |