

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
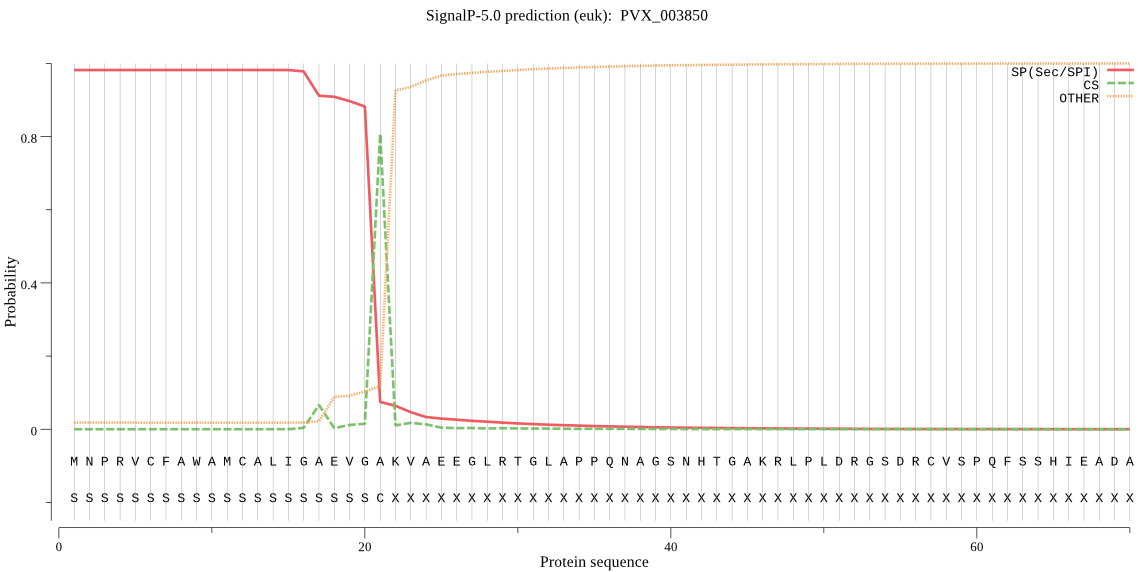
| PVX_003850 | SP | 0.005445 | 0.994533 | 0.000022 | CS pos: 21-22. VGA-KV. Pr: 0.8497 |
MNPRVCFAWAMCALIGAEVGAKVAEEGLRTGLAPPQNAGSNHTGAKRLPLDRGSDRCVSP QFSSHIEADAPPRSADDAAGRGQPPQQVEHPPNRVTNVMAPPMSKQSAVEVRSALLKNHD GVKITGTCNAKVQLFLVPHISISVEAESNTIQLGRKLEDVTITKEFYRGVGGKSSPLLQF EEDADSLLNQCAEGKTFKFVVVIRGEELILKWKVYEKVPSPSDNNKVDVRKYLLKNLGRP ITAIQVHSGKEESDVFLLESKAYLLRQDIPRTCERIASSCFLSGNVDIEGCYKCTLLSED TKLGSPCFSYLPPEVKHNYEKVKMKAQDEGDLGEAQLEELIRRILHAVQRKGKSNPPRKG ATEEDYPNDNLRELLLRYCQVMKKVDRSGTLEEHEVGNEMDVLANLEGLLRKHPKEEIPA LREKLKNPAICMKDAAKWVEQKRGLALPLFEYKHLQRRVTTAVDSQESNSADKENIIKGE HIAAYRAVIDLSQRDQMNHSNLSGEMFCNEDYCDRWKDKSSCFSSIETEEQGNCNLSWLF TSKTHLETIRCMKGYDHLGSSALYVASCSKRGKKSRCVSGSNPYEFLTIVEESGFLPPAV WLPYSYGDVGNGCPKREDHWHNLWEGVKLLEPADEPNSVSTKGYTSYESDDFRGDIQTFV NMVKSQVRSKGSVMAYIKVLDILSYDFNGKEVHSLCGDKRPDHAVNIIGYGNYVTPEGVK KSYWLMRNSWGKYWGDGGNFKVDTHGPPGCQHNFIHTAAVFNLSMPMAEGPSKGEAHLYD YYLKSSPDFLGNIYHKISGRRSGAVEKGRTTGNQSLTVQGQEGWSDLLEGTTPSTTANPN PVGELKGAQDVPKRGNTGKRISEDPQEEVLNEELIPQLPLAPISQAAATLGEGQVEATPL PPLTVAASLPLRGDVLLERDGTNEGSKEAATVEVMHILKYIKNGKVKLGIATYRDDSDIA GDHSCSRSLAQNSEQLAECIQFCHDEWPNCRGEASPGYCLAQRRRTGDCFFCYV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_003850 | 248 S | IQVHSGKEE | 0.996 | unsp | PVX_003850 | 248 S | IQVHSGKEE | 0.996 | unsp | PVX_003850 | 248 S | IQVHSGKEE | 0.996 | unsp | PVX_003850 | 465 S | TAVDSQESN | 0.994 | unsp | PVX_003850 | 492 S | VIDLSQRDQ | 0.995 | unsp | PVX_003850 | 524 S | SSCFSSIET | 0.995 | unsp | PVX_003850 | 646 S | KGYTSYESD | 0.997 | unsp | PVX_003850 | 798 S | YHKISGRRS | 0.991 | unsp | PVX_003850 | 862 S | GKRISEDPQ | 0.996 | unsp | PVX_003850 | 957 S | YRDDSDIAG | 0.997 | unsp | PVX_003850 | 74 S | APPRSADDA | 0.991 | unsp | PVX_003850 | 220 S | EKVPSPSDN | 0.996 | unsp |