

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
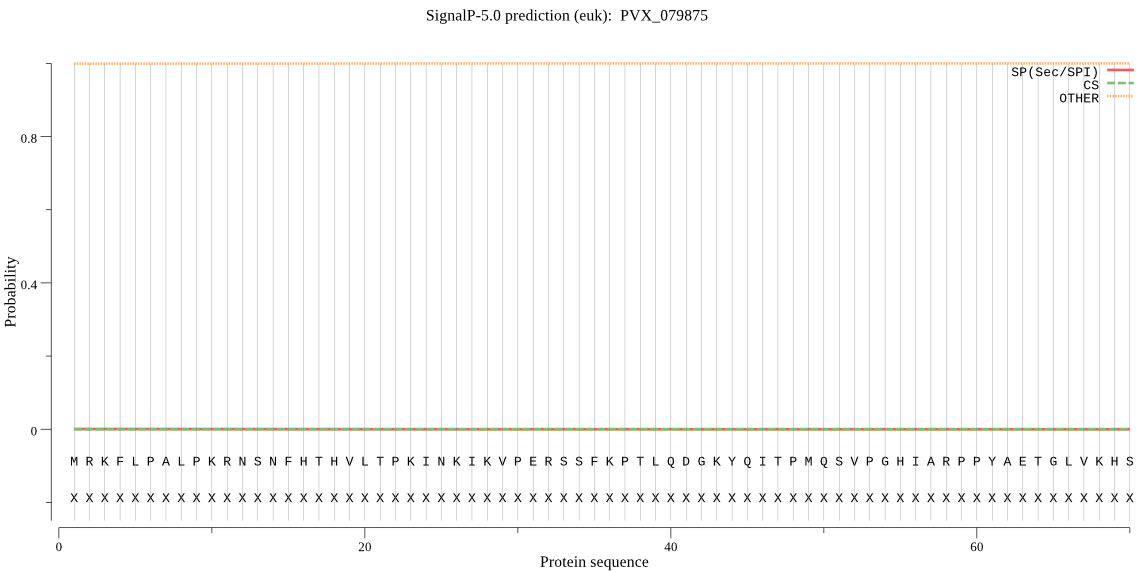
| PVX_079875 | OTHER | 0.759543 | 0.001120 | 0.239336 |
MRKFLPALPKRNSNFHTHVLTPKINKIKVPERSSFKPTLQDGKYQITPMQSVPGHIARPP YAETGLVKHSNVDYEIKDEESIAKMKEAAKIAAQCLKLCLENSKAGVTTDEIDKMAFNFY VEKGAYPAGINFHGFPKTVCASPNEVVCHGIPNLRKLKDGDIITYDCTVYIDGVFGDCAG TTGIGTISKSHQKLVDVSKECLYKAISVCKHGQKFSEIGRVITEHATRNGFNVIQEFCGH FIGRNMHMYPLIEHHYPNGHPDDEYMQVGQIFTIEPILSEGSRKIHTWKDQWTVCTNDNA FCSQWEHTILVQQNGAEILTQCD
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PVX_079875 | 34 S | PERSSFKPT | 0.994 | unsp | PVX_079875 | 265 Y | PDDEYMQVG | 0.99 | unsp |