

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_080490 | OTHER | 0.881589 | 0.000362 | 0.118048 |
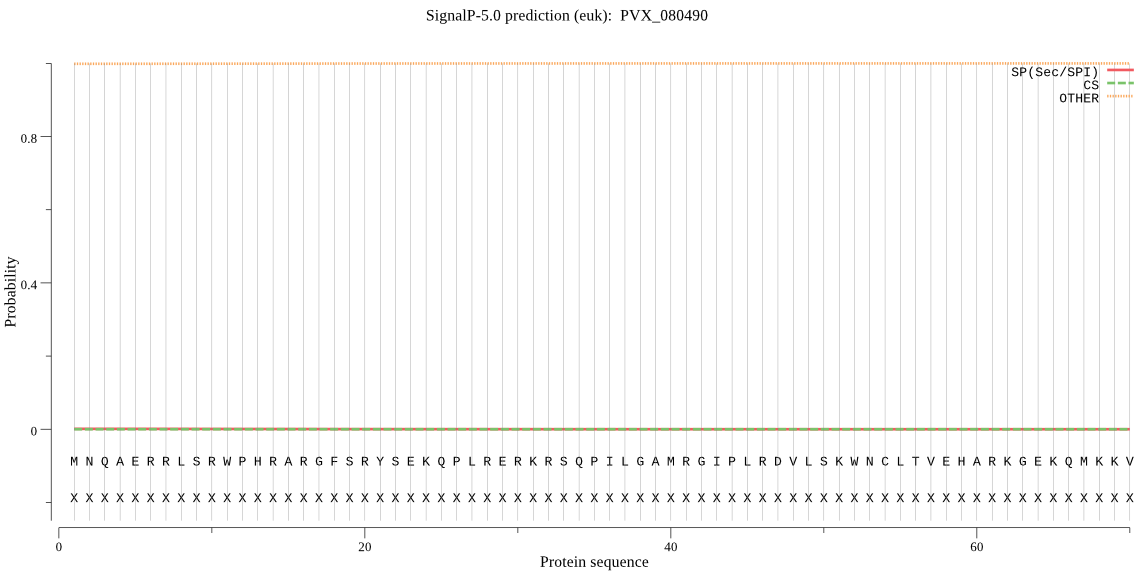
MNQAERRLSRWPHRARGFSRYSEKQPLRERKRSQPILGAMRGIPLRDVLSKWNCLTVEHA RKGEKQMKKVIKKTYAELHVKYEEVRAKYKDTFPTWRDKRMKVLKRVGPFFQKGVFIRSI PIDGGQMKTILQKTFSLGGRVFFKKLKLLYRDANVPYYKNLFYHHFGRAPVTYTLMSLHI LVFLLWVKAKPGDTYSYIGMPPRGYYYPHSNAYTPLGGRNASSNYSSPFRLLNFLTMEFM YTHFCCGVQQLRERRLYTLVTNLISHNTGQSLLLNTISLFYIGRSFEMAINSRNFFLTYF LSGIISSYVQICYHKGGRFFLSPTSSSPYGNVCVLGASGSISSILATYTLMFPRSSIYLY GVLALPLALFTSLYGANEVYCVLTDKKDNTGHVAHLTGMFLGILYYYAYVKGRVPL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_080490 | 22 S | FSRYSEKQP | 0.995 | unsp | PVX_080490 | 22 S | FSRYSEKQP | 0.995 | unsp | PVX_080490 | 22 S | FSRYSEKQP | 0.995 | unsp | PVX_080490 | 9 S | ERRLSRWPH | 0.996 | unsp | PVX_080490 | 19 S | ARGFSRYSE | 0.994 | unsp |