

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
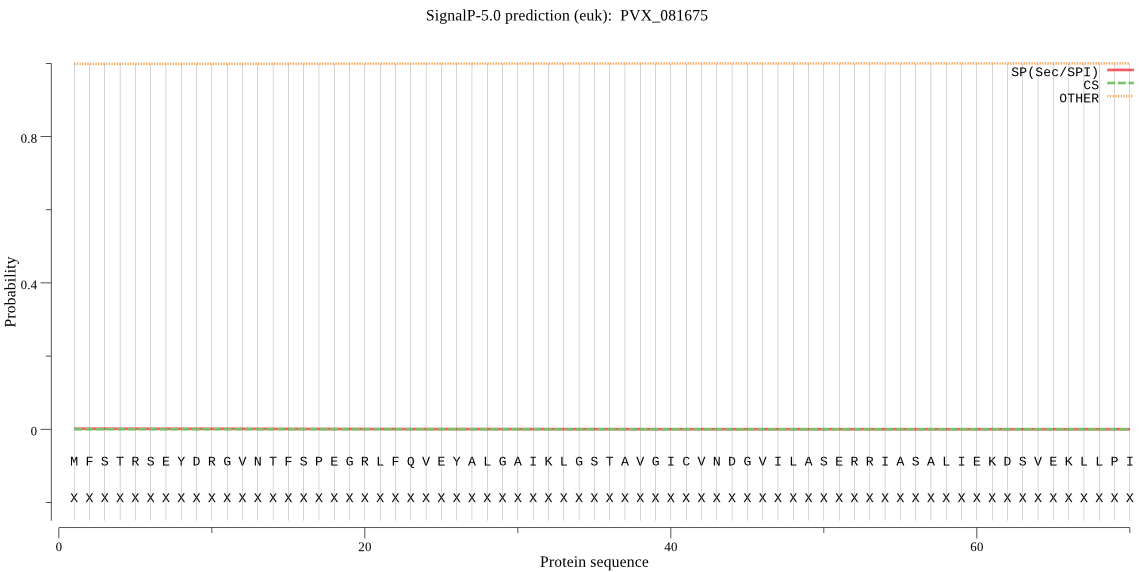
| PVX_081675 | OTHER | 0.999312 | 0.000154 | 0.000534 |
MFSTRSEYDRGVNTFSPEGRLFQVEYALGAIKLGSTAVGICVNDGVILASERRIASALIE KDSVEKLLPIDDHIGCAMSGLMADARTLIDYARVECNHYKFIYNENINIKSCVELISELA LDFSNLSDNKRKKIMSRPFGVALLIGGVDKNGPCLWYTEPSGTNTRFLAASIGSAQEGAE LLLQENYNKDMTFEEAEILALTVLRQVMEDKLSSSNVEIAAVKKSDQTFYKYKTEDISRI IEALPSPIYPTIDMTA
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_081675 | T | 255 | 0.632 | 0.041 | PVX_081675 | T | 251 | 0.563 | 0.315 | PVX_081675 | T | 4 | 0.532 | 0.030 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_081675 | T | 255 | 0.632 | 0.041 | PVX_081675 | T | 251 | 0.563 | 0.315 | PVX_081675 | T | 4 | 0.532 | 0.030 |