

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
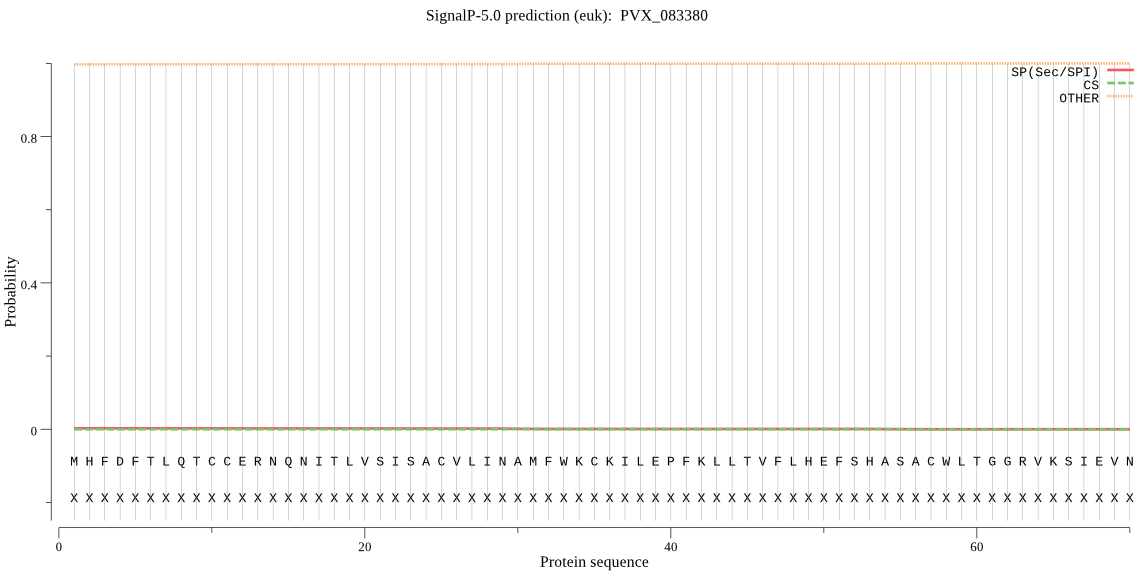
| PVX_083380 | OTHER | 0.999968 | 0.000014 | 0.000019 |
MHFDFTLQTCCERNQNITLVSISACVLINAMFWKCKILEPFKLLTVFLHEFSHASACWLT GGRVKSIEVNRNHGGCTNTIGGNKFLILPAGYIGSCFYGMFFILMAYINKWTLITSAAFL CFLLLIVLIFYANNFFLRLLCVLFLTLTISVWVLCVHFKEDVQYWPLKIIMTFIGVLNEI YSMVDIIEDLITRSVPESDAYKYAELTKCNSKFCGALWFVVNLTFICLTVYLIGAIHVKS FDD
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PVX_083380 | 66 S | GRVKSIEVN | 0.993 | unsp |