

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
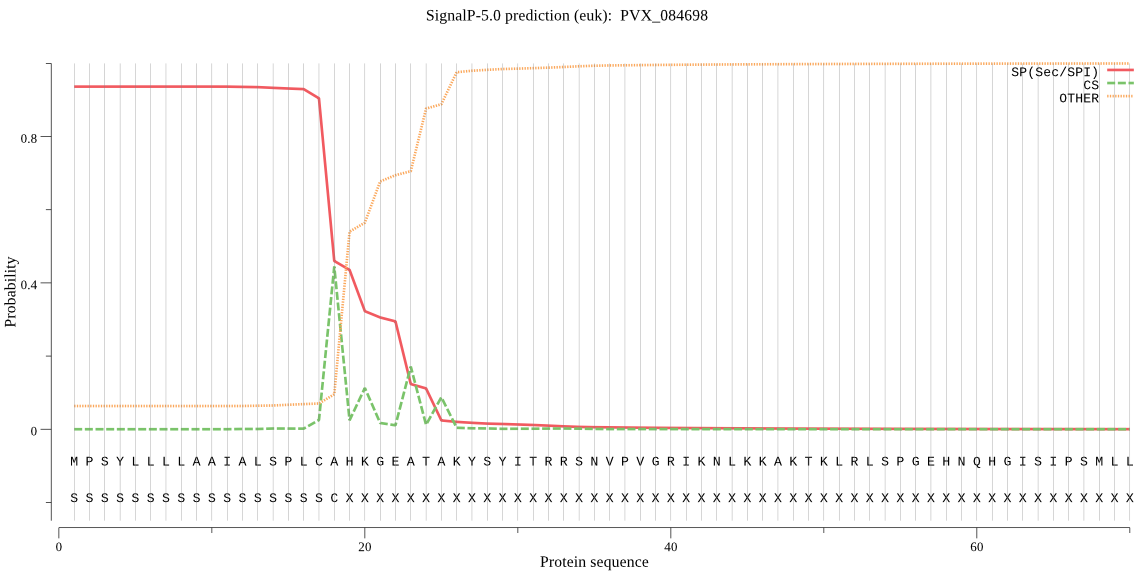
| PVX_084698 | SP | 0.007605 | 0.990727 | 0.001668 | CS pos: 23-24. GEA-TA. Pr: 0.5998 |
MPSYLLLLAAIALSPLCAHKGEATAKYSYITRRSNVPVGRIKNLKKAKTKLRLSPGEHNQ HGISIPSMLLSKRIIFLSSPVYPHISEQIISQLLYLEYESKRKPIHLYINSTGDLENNKI INLNGMTDVISIIDVIDYISSDVYTYCLGKAYGISCILASSGKKGFRFSLKNSSFCLNQS YSVIPFNQATNIEIQNKEIMNTKRKVVEIIANNTGKEKSHIERILERDRYFSAPEAVQFN LVDHILERE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_084698 | 232 S | DRYFSAPEA | 0.996 | unsp | PVX_084698 | 232 S | DRYFSAPEA | 0.996 | unsp | PVX_084698 | 232 S | DRYFSAPEA | 0.996 | unsp | PVX_084698 | 54 S | KLRLSPGEH | 0.998 | unsp | PVX_084698 | 169 S | GFRFSLKNS | 0.996 | unsp |