-
-
-
-
-
Computed_GO_Process_IDs: GO:0006465
-
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
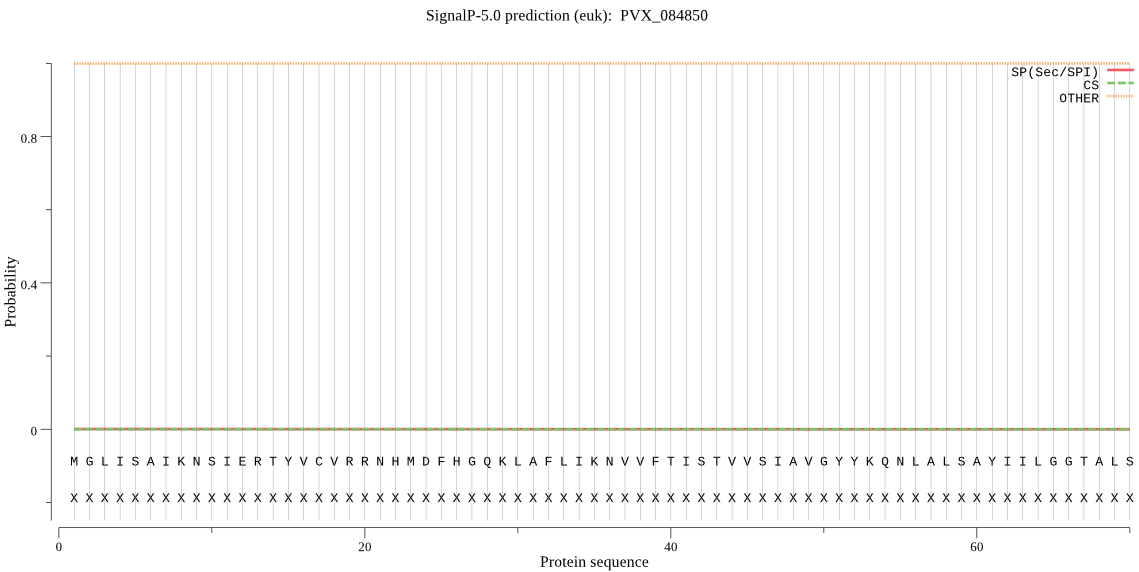
>PVX_084850
MGLISAIKNSIERTYVCVRRNHMDFHGQKLAFLIKNVVFTISTVVSIAVGYYKQNLALSA
YIILGGTALSALLIMPTWPIYNKHNIQWESSEDPMEDRKRR
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/PVX_084850.fa
Sequence name : PVX_084850
Sequence length : 101
VALUES OF COMPUTED PARAMETERS
Coef20 : 4.688
CoefTot : -0.010
ChDiff : 5
ZoneTo : 11
KR : 1
DE : 0
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.988 1.971 0.379 0.728
MesoH : 0.136 0.734 -0.215 0.340
MuHd_075 : 26.731 13.243 6.192 5.519
MuHd_095 : 26.257 19.071 7.965 6.919
MuHd_100 : 25.834 20.779 8.612 6.404
MuHd_105 : 24.342 20.030 8.459 5.221
Hmax_075 : 10.033 15.600 2.810 5.030
Hmax_095 : 15.200 19.500 3.803 6.610
Hmax_100 : 16.500 20.000 4.890 6.610
Hmax_105 : 18.500 21.400 5.493 6.180
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.4428 0.5572
DFMC : 0.6173 0.3827
-
Fasta :-
>PVX_084850
AGAAGCCAACCGGTAGGCTAACAAATAAAGAGGTTATCATTCACAACAGGGGGGAGTTAT
AAACTTTACGCGAAGCGAATTGTACTGGAGGAGTCCCCCTTTTAGGCTCCCCCTTTATGT
CACCGCCATCTTTGCGATGAGATATGCCCTTGCATGGCAACTCGCATACGCACGTACAGC
TACGAAGGAGGACCAATGCACAGCAGGAGGATTCCCCCTCACCTCTTCACCATTAATAGA
ACAGTTCATCCGAGTGAAATAGAATAACCCCCCATTCGCGCCGCTCTCACGGAGAGTATA
CATCCCTGCTTACGTTCCCCCGTGCAACGCTGCGAGTGGGTAACGAAGAAGAAAAGGGGA
AAAACCTCCTCGAAAGTCCTGCCCCCCTCCTCCTCCCCCATTCGAACGATCACCCCATGG
GCCTAATTAGCGCGATTAAGAACAGTATAGAGCGCACCTACGTGTGTGTCCGGAGGAATC
ACATGGACTTCCACGGCCAGAAGCTCGCATTCCTAATCAAAAACGTCGTCTTCACCATCA
GCACGGTAGTCTCCATTGCAGTCGGTTATTACAAGCAGAACTTAGCCTTGAGTGCCTACA
TCATTTTGGGCGGCACCGCCTTGTCAGCACTGCTAATCATGCCCACGTGGCCCATATACA
ACAAGCACAACATTCAGTGGGAAAGTTCAGAAGATCCCATGGAGGATAGGAAGCGAAGAT
AA
- Download Fasta


