

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
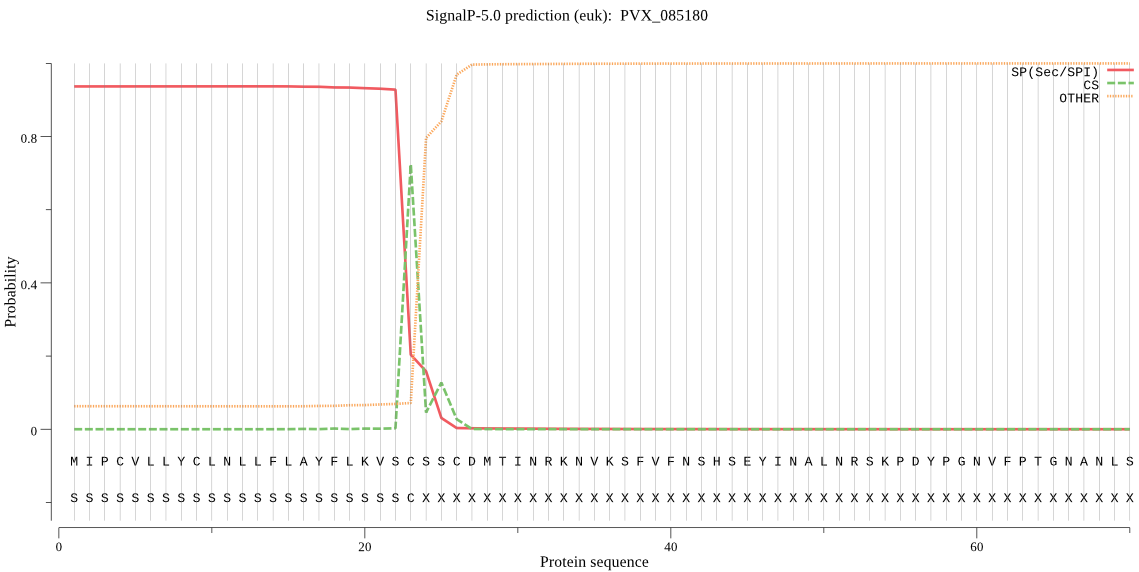
| PVX_085180 | SP | 0.275215 | 0.723366 | 0.001419 | CS pos: 23-24. VSC-SS. Pr: 0.7282 |
MIPCVLLYCLNLLFLAYFLKVSCSSCDMTINRKNVKSFVFNSHSEYINALNRSKPDYPGN VFPTGNANLSKAGGHAAAGAEGAVKAGKGGKKKQEEKKPEGKKPEEKKVEEKKPAERKPQ EKKPEEKKPAEKKPAERKPQEKKPEEKKPAEKKSAEKKLVETKPEEKKVEGKKPEEKKVE EKKPAEKKPAERKPQEKKPEEKKPAEKKSAEKKLVEKKPQEKKAEQKEAVKPEKKEEKRL QKEEQKVVKKLGQKLGPKLGSQVEPKLQPTVGPKLGSQIEPKLQATIGPKLGFPVDPKLQ PTVGPKLGFPVEPKLQPRVVSKLGSSMEPKLQPTVGPKLGSQIEPKLQATIGPKLGFPVE PKLQPRVVSKLGSSMEPKLQPKVVPKLGSSMEPKLQPKVVPKLGSSMEPKLQPKVGPKLG SSMEPKLQPKVGPKVGKKVQSRTHQKTKEDHAQSQKVGDPVEAVPMPNQNVDEVVKENLL PSEKVDQWIQSIPLPNQKDGANSSLPNEKVNQWIQAFPLSNERVDKKPPTERPTDKLATR TPLSERRRAKKRDEWIKVMPLSNEKVDHRVQADLLSDEAANRTTLSERILSQVETQRIPS TRLPTQVDNPRVQSECAQGKIKMANQKTPGKRVPSERVGQKKPKEDIATPPEVRGIKKYA DKLQRIAKREKREVAKEEKREVAKEEKREVAKEEKKVETNEETKGDSTEDTSGDTSGDTS EDAHTDSYEGVPQGALKEQKWSDKKERNKKGWFGTYWSQSTKKSKPSKPANAEKKGAKKS NLIPLICKRANMRKDEEFKLTEENCLVTELPKKDLDASEKKNLENEPPLDKEGMIQKVDG SYLRVQPIDYNRAEKGIKDEEKITRRGSFGFLTSPSSSYHLGEGIINTQEPLGVCDAEAG KHLDILSGQVVRDGEGVDPCNAIKRSDGDVVSARGEHNVIFGGVDQVKRRGAKSKRRREG KRRRGEAKRKAGGARKEEAKREEEEEEEVKVGEDEAKKEEEEEVKVGEDEAKKEEEEEEE EEVKTGEDYTKKEKEEEEEEEEAKVGDDEAKKEEEEEEEVKMGDEETKEEKEEKEEKEEE EAKVEDEDGNVKRKDDTEDSGIDSDDAQKKHAEEVPGEPAEGAHLEDKLYDDYEDEDDYD DDEDDYDEEEEEEEEEDDEEEEEEEEEEEEEEEADDIDEVINRHLEEVEKIEQSLGLAKK EGSSEMVLVPTPSKGSKFDNTDTESVASASTSTSGGNDDVGGEEKEFGKKDRFRYKSTNV NVYERYKKDLKLYSEMVKDNELNEFYKEIIRNRELVDVNYKNEYALKMKYHKGYDFSLHN LHIVNEQKASEKFYSNFVATCDEEDNATFQDVNYFADLTKVDIDPVMFKEFYIRSSVCDG EYLPLKSTLVEKICFLSNHLKKNPYINRIKIKSKKSILYRLFLNIVKTFKKLSLKQCIKN VEESNLLDILFLLNEEIVKESKKDLNIGKCLNNVIARNRDWYDLINFIVTIMFGCTTYMK EHYFLTEEERNALITTDQVVDLPIFEYLNSNVIDIFVPANVQTQIDVDLLKDITDAPEDK KKMYQTWYVCYLFTHKVYLVNKMWNVIKKWEASKFIPNPWYIDHIGSALVPQIMNLHHLE EDKYVFFRYIDSLQIFVSFKRSKKYPLPNYDKNNFHLVENIVAFQGTASPFMWILNVIYE LTSYPFLTKGKLHKAYLFIFKRVVKPYLHVLKKNILNEIKDEKSKYTKENPYVIIFTGHS FGAAIAHLSSFYLAKILKARNNPKVKVLSVTFGMPMFFDEQFSEDFRKSGVISNNISIDY DPIPCTMAIPGFNDFKDPDEEKKLSIILKVNDLKSINHDFGENIFNGNELFHHERNQPRS MIHLLTKYIFNNNIFLELGELHISQTHYFFYYVFLTLLSGWANEAEWGTFFLIPFFLFDI INFTHSEIMTKSKEVHKKYREDLMKKLQSSQKNKGGGGKK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_085180 | 591 S | ERILSQVET | 0.994 | unsp | PVX_085180 | 591 S | ERILSQVET | 0.994 | unsp | PVX_085180 | 591 S | ERILSQVET | 0.994 | unsp | PVX_085180 | 635 S | KRVPSERVG | 0.991 | unsp | PVX_085180 | 707 S | TKGDSTEDT | 0.998 | unsp | PVX_085180 | 760 S | YWSQSTKKS | 0.993 | unsp | PVX_085180 | 868 S | TRRGSFGFL | 0.994 | unsp | PVX_085180 | 954 S | RGAKSKRRR | 0.997 | unsp | PVX_085180 | 1100 S | DTEDSGIDS | 0.997 | unsp | PVX_085180 | 1139 Y | DEDDYDDDE | 0.992 | unsp | PVX_085180 | 1216 S | PSKGSKFDN | 0.994 | unsp | PVX_085180 | 1376 S | YIRSSVCDG | 0.998 | unsp | PVX_085180 | 1461 S | IVKESKKDL | 0.996 | unsp | PVX_085180 | 1624 Y | EEDKYVFFR | 0.992 | unsp | PVX_085180 | 1950 S | KLQSSQKNK | 0.992 | unsp | PVX_085180 | 454 S | DHAQSQKVG | 0.991 | unsp | PVX_085180 | 544 S | RTPLSERRR | 0.993 | unsp |