

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
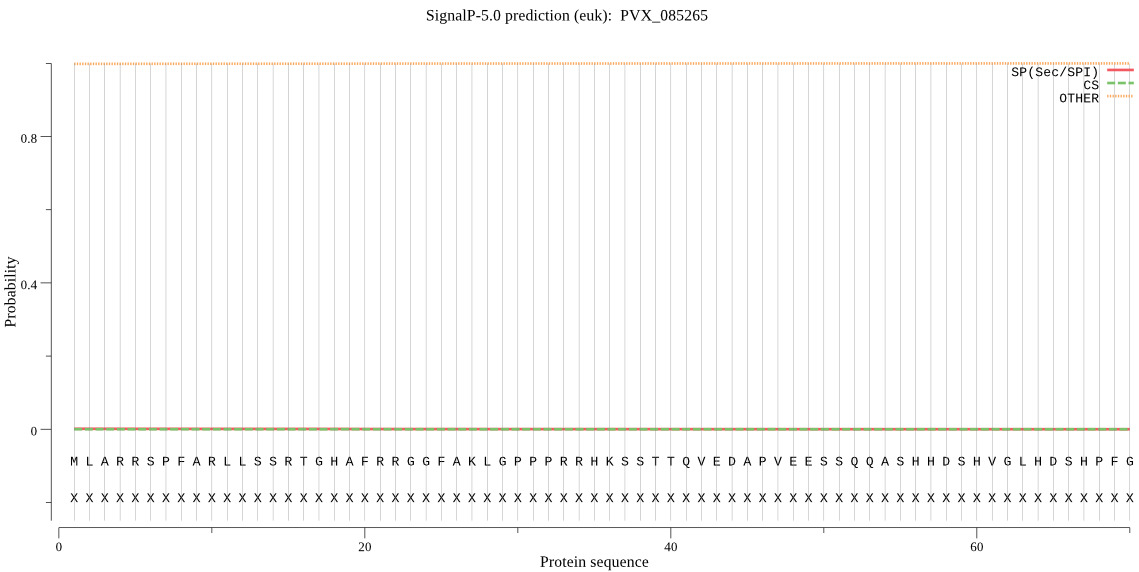
| PVX_085265 | mTP | 0.014601 | 0.000029 | 0.985369 | CS pos: 35-36. RRH-KS. Pr: 0.2784 |
MLARRSPFARLLSSRTGHAFRRGGFAKLGPPPRRHKSSTTQVEDAPVEESSQQASHHDSH VGLHDSHPFGHRFAHHKYVDFFRDESMSVGIVSFRSAVHGKKNILGDFLEELKNVVEHIT NVMANEETNTFYIKEFPRGDNYLVQNMRNRIPYVDNRLKVLILTGGNEPIGSGEASMSGS TPDYNSFLKNDEQLNVQLANSFRYLCNTIQHLPLITISSINGQCHNSGMDLLLSTDFKLS SEESTFGFNKAHLGLLPYGGGTQKLFRAIPMSYAKHLLLTGDTIRAKDALRINLIDICMG HNEDHFVQNSCVHFDANLAKKEKMQIIKESVGTFFADVFSNELFKDRVKDSCFVFSLFFA FQFLFIPTNALQNVKMSILEGMSLAEPNLYLDHDRAVFERNINAPQRAEILAYLKRRVSG APSVPT
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_085265 | T | 426 | 0.600 | 0.407 | PVX_085265 | T | 40 | 0.579 | 0.131 | PVX_085265 | T | 39 | 0.563 | 0.061 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_085265 | T | 426 | 0.600 | 0.407 | PVX_085265 | T | 40 | 0.579 | 0.131 | PVX_085265 | T | 39 | 0.563 | 0.061 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_085265 | 419 S | KRRVSGAPS | 0.994 | unsp | PVX_085265 | 419 S | KRRVSGAPS | 0.994 | unsp | PVX_085265 | 419 S | KRRVSGAPS | 0.994 | unsp | PVX_085265 | 55 S | SQQASHHDS | 0.995 | unsp | PVX_085265 | 240 S | DFKLSSEES | 0.997 | unsp |