

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
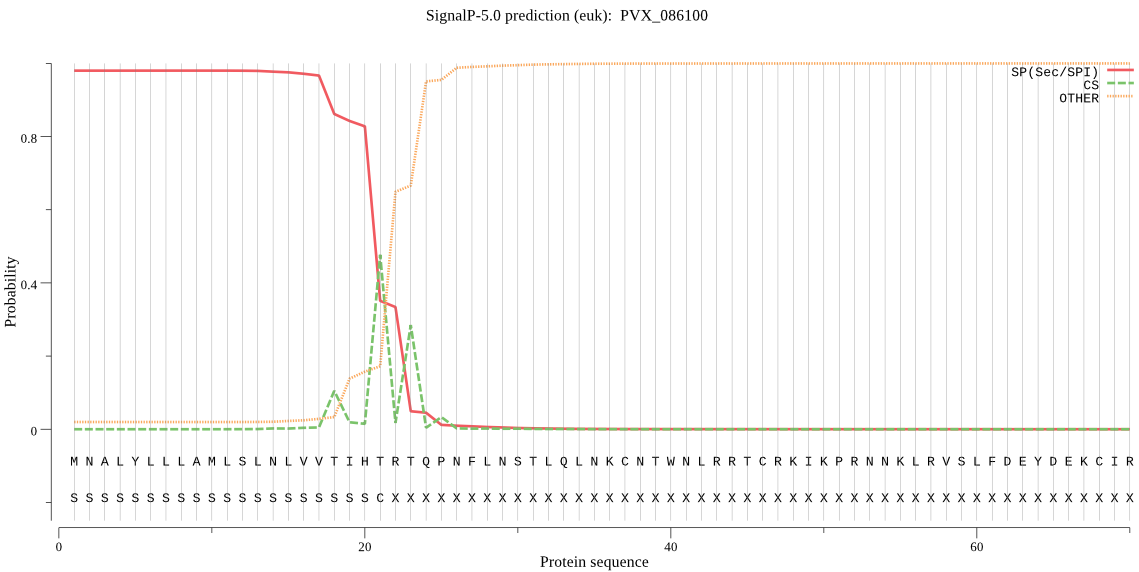
| PVX_086100 | SP | 0.028111 | 0.969266 | 0.002623 | CS pos: 21-22. IHT-RT. Pr: 0.7048 |
MNALYLLLAMLSLNLVVTIHTRTQPNFLNSTLQLNKCNTWNLRRTCRKIKPRNNKLRVSL FDEYDEKCIRALIMAREVAKNHNEKEILLKHLLIAIIRIDSNLVKHILNFCNISLTTFLD KLNNEMNKTGGETNFGTNRDEPNASPGEVPRQLGMPTDQHHPPDNSREVVEGGAYPANDP NGENQLMSTFINKHIQDIEDKINQLKNLEEGQSDISTEGGDADSSVGGADERDEAASNLI RDAAKRVLGGPPGESPSELPSELPSDVSKELSRQLPSDLQGDPPDEKADERADDKADIKF SQNCKRVLHNAVLEARRKKKMFVSVTDILIALINMAQESQNCEFLRYLSELNISVNDLKR ELTGYDEGNCPTPPTGTSHRSNAQRDGDLNANSAEKRGGGDSQSGTPEKRDDNQRMRSLN NRLNNEPPNHHLMNNLNSDYLHQTREFKNYEESSFPPNGKLFNSAYVKDCLTDMVQAAYE KGDEHFFGRKKEIKRIIEILGRRKKSNPLLIGESGVGKTAIIEHLSHLILKDKVPYHLRN CRIYQLNVGNIVAGTKYRGEFEEKMKHLLSNMNKKKKNILFIDEIHVIVGAGSGEGSLDA SNLLKPFLSSDNLQCIGTTTFQEYTKYIESDKALRRRFNCVPVKPFTGKETLLLLKKIKY SYEKYHNIYYTNEALKSIVMLTEDYLPTANFPDKAIDILDEAGAYQKIRYEVFMRRRLRG ENHERSEWGERVERGGQAKSGQENGGQVNGGQANGGKLRSDQPSGDEHGSADQDGIANQD SGGDARSYLQNMHIKYVTSDVIENIVSKKASISYIKKNKKEEEKIIKLKEKLNKIIIGQE KVIDILSRYLFKAITNIKDPNKPIGTLLLCGSSGVGKTLCAQVISKYLFNEDNLIVINMS EYIDKHSVSKLFGSYPGYVGYKEGGELTESVKKKPFSIILFDEIEKAHGEVLHVLLQILD NGMLTDSKGNKVSFKNTFIFMTTNVGSDIVTDYFKLYNRNYSNMGFKYYLSKRPNRGEAD GSLLPEGVNSVANSVEEDPTGLSSNSGGDLQTNRKDDHPVEKSPTQQPNSYDLFEEKLRT NKWYEELQPEIEEELKKKFLPEFLNRIDEKIIFRQFLKRDVINIFQNMIDDLKKRIKKRK NLNLIIEPDVIKYICSDENNIYDMNFGARSIRRALYRYIEDPIASFLISNLCEPNDSIYV SLSSGNKIDVQLVKAPVEQLVL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_086100 | 216 S | QSDISTEGG | 0.996 | unsp | PVX_086100 | 216 S | QSDISTEGG | 0.996 | unsp | PVX_086100 | 216 S | QSDISTEGG | 0.996 | unsp | PVX_086100 | 255 S | PPGESPSEL | 0.995 | unsp | PVX_086100 | 764 S | SDQPSGDEH | 0.995 | unsp | PVX_086100 | 781 S | ANQDSGGDA | 0.996 | unsp | PVX_086100 | 1034 S | SVANSVEED | 0.996 | unsp | PVX_086100 | 59 S | KLRVSLFDE | 0.994 | unsp | PVX_086100 | 145 S | EPNASPGEV | 0.991 | unsp |