

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
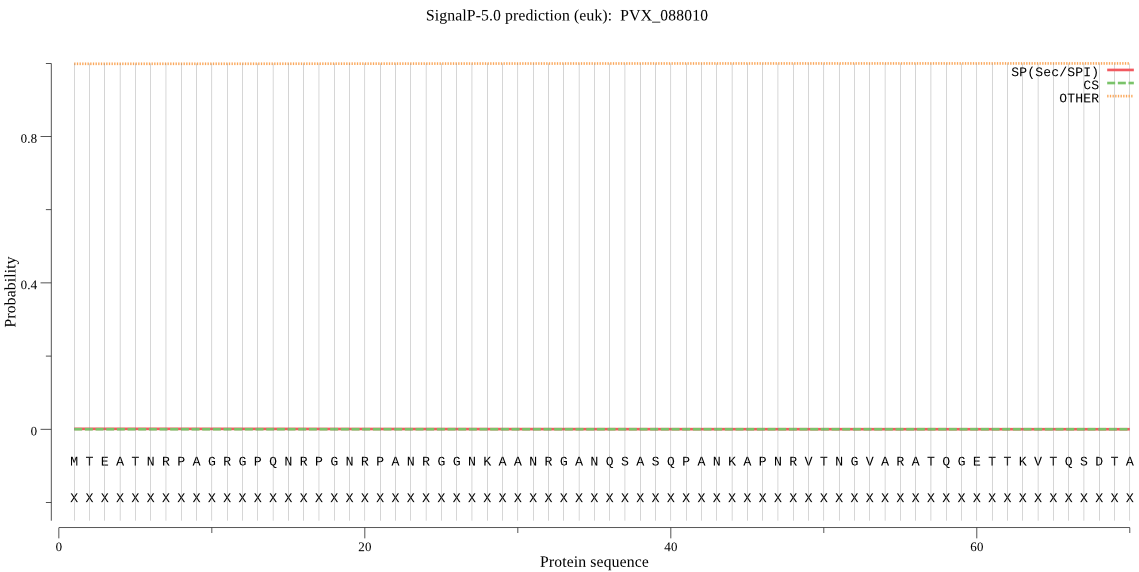
| PVX_088010 | OTHER | 0.999998 | 0.000001 | 0.000001 |
MTEATNRPAGRGPQNRPGNRPANRGGNKAANRGANQSASQPANKAPNRVTNGVARATQGE TTKVTQSDTAKVTAKESEKEAMNNTANQAGNKTATEPERKASPEPEKKAPSEPEKKVPIE PDRKVSSEPERKISSEPERKAVTEPERKISSEPERKASTEAERKASTEAERKISSEPERK ASTEAEKRTSPEPEKKTSNEPERKVSTEPEKKVSIEPEKKVSIEPDKKAPTEPERKAPTE PERKVPAEPEKKAPTDAERKVSTEPEKATGQPAKQAISGGAKEAINQTEKGAKETPNRDA NKAAIAASAPATSAPATSAAATSAAAAAAPDDGGSQSGALKTGSFVSRDGLELKTYEWVV NKPVGIILLVHALNSHVRFEYLKHNVIIESKEKATLLDGKNYYIYKDSWIEQLNKNGYSV YGLDMRGHGQSGCVQNVKTHINDFHDLVYDVLEYANIVYDSLCTGGKKKKKKSPSGGVNT SSDTAPSGGLSSSASSSDDDDTSQGSRTSSGNAPPSGANSVRRSDANSMKRSDANNMRRS DANNMRRSDANNMRRSDVPPFFLMGLSMGGNIVLRILELGGKKGDESIKRLNIKGVISLA GMISLDDLKKKPEYKYFYIPMGKLASMVLPTMRMTPSLNFKMFPFINDLFSFDAHCYPKP VTNRFGSELLKAVDALRNDVKFIPEEAQILLVHSVLDSACSYNGVLKFFKSIQTKNKELF TIEDMDHILPLEPGNERILKKVIDWLAQLQCVRG
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_088010 | T | 322 | 0.647 | 0.262 | PVX_088010 | T | 239 | 0.601 | 0.400 | PVX_088010 | T | 502 | 0.598 | 0.031 | PVX_088010 | T | 317 | 0.587 | 0.229 | PVX_088010 | T | 2 | 0.571 | 0.400 | PVX_088010 | T | 508 | 0.567 | 0.039 | PVX_088010 | T | 231 | 0.565 | 0.433 | PVX_088010 | T | 189 | 0.560 | 0.032 | PVX_088010 | T | 255 | 0.552 | 0.284 | PVX_088010 | T | 197 | 0.545 | 0.057 | PVX_088010 | T | 183 | 0.535 | 0.064 | PVX_088010 | S | 323 | 0.525 | 0.092 | PVX_088010 | T | 312 | 0.522 | 0.462 | PVX_088010 | S | 495 | 0.521 | 0.047 | PVX_088010 | S | 506 | 0.517 | 0.021 | PVX_088010 | T | 263 | 0.515 | 0.067 | PVX_088010 | T | 484 | 0.504 | 0.041 | PVX_088010 | T | 5 | 0.500 | 0.158 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_088010 | T | 322 | 0.647 | 0.262 | PVX_088010 | T | 239 | 0.601 | 0.400 | PVX_088010 | T | 502 | 0.598 | 0.031 | PVX_088010 | T | 317 | 0.587 | 0.229 | PVX_088010 | T | 2 | 0.571 | 0.400 | PVX_088010 | T | 508 | 0.567 | 0.039 | PVX_088010 | T | 231 | 0.565 | 0.433 | PVX_088010 | T | 189 | 0.560 | 0.032 | PVX_088010 | T | 255 | 0.552 | 0.284 | PVX_088010 | T | 197 | 0.545 | 0.057 | PVX_088010 | T | 183 | 0.535 | 0.064 | PVX_088010 | S | 323 | 0.525 | 0.092 | PVX_088010 | T | 312 | 0.522 | 0.462 | PVX_088010 | S | 495 | 0.521 | 0.047 | PVX_088010 | S | 506 | 0.517 | 0.021 | PVX_088010 | T | 263 | 0.515 | 0.067 | PVX_088010 | T | 484 | 0.504 | 0.041 | PVX_088010 | T | 5 | 0.500 | 0.158 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_088010 | 111 S | KKAPSEPEK | 0.994 | unsp | PVX_088010 | 111 S | KKAPSEPEK | 0.994 | unsp | PVX_088010 | 111 S | KKAPSEPEK | 0.994 | unsp | PVX_088010 | 126 S | DRKVSSEPE | 0.996 | unsp | PVX_088010 | 134 S | ERKISSEPE | 0.996 | unsp | PVX_088010 | 150 S | ERKISSEPE | 0.996 | unsp | PVX_088010 | 158 S | ERKASTEAE | 0.997 | unsp | PVX_088010 | 166 S | ERKASTEAE | 0.997 | unsp | PVX_088010 | 174 S | ERKISSEPE | 0.996 | unsp | PVX_088010 | 182 S | ERKASTEAE | 0.997 | unsp | PVX_088010 | 190 S | EKRTSPEPE | 0.998 | unsp | PVX_088010 | 198 S | EKKTSNEPE | 0.992 | unsp | PVX_088010 | 206 S | ERKVSTEPE | 0.997 | unsp | PVX_088010 | 214 S | EKKVSIEPE | 0.993 | unsp | PVX_088010 | 222 S | EKKVSIEPD | 0.993 | unsp | PVX_088010 | 262 S | ERKVSTEPE | 0.997 | unsp | PVX_088010 | 473 S | KKKKSPSGG | 0.997 | unsp | PVX_088010 | 495 S | SSSASSSDD | 0.996 | unsp | PVX_088010 | 496 S | SSASSSDDD | 0.998 | unsp | PVX_088010 | 497 S | SASSSDDDD | 0.996 | unsp | PVX_088010 | 509 S | GSRTSSGNA | 0.99 | unsp | PVX_088010 | 520 S | SGANSVRRS | 0.996 | unsp | PVX_088010 | 528 S | SDANSMKRS | 0.998 | unsp | PVX_088010 | 587 S | KGDESIKRL | 0.992 | unsp | PVX_088010 | 77 S | TAKESEKEA | 0.995 | unsp | PVX_088010 | 102 S | ERKASPEPE | 0.998 | unsp |