

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
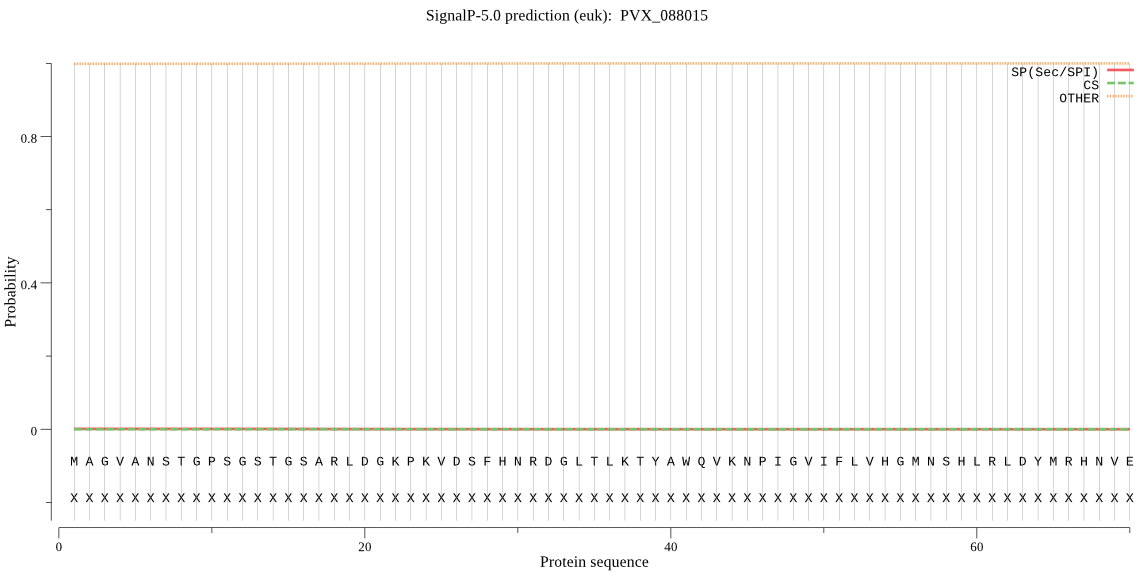
| PVX_088015 | OTHER | 0.999860 | 0.000108 | 0.000031 |
MAGVANSTGPSGSTGSARLDGKPKVDSFHNRDGLTLKTYAWQVKNPIGVIFLVHGMNSHL RLDYMRHNVEIVSPEKAILKDADNYYVYKNSWVEQLNKSGYSVYGLDLQGHGQSDGWRNL KTNMKKFDDLVYDLLQYINRVHDVICLTGRKDAKGGDAGKSGSAVASPASPTPAAGAASP SRSIHSNLKNAKTPPFYLMGLSMGGNIVLRVLEILGKSKTHSEKLNIRGCICLAGMISID ELASRASYKFLFIPFSKFFATVFPSLRLTPSLYFKKYPYVNQIYSYDRNRNKKSITCKMG YELLNAIENLNNDMQHIPKDIPILFVHSRHDGTCFFGGVETFFKKINTDVKELHVLDDMD HVLTMEPGNERVLQKVLAWLAARG
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_088015 | T | 172 | 0.671 | 0.674 | PVX_088015 | S | 170 | 0.571 | 0.178 | PVX_088015 | T | 8 | 0.566 | 0.219 | PVX_088015 | S | 181 | 0.547 | 0.269 | PVX_088015 | S | 167 | 0.539 | 0.230 | PVX_088015 | S | 179 | 0.535 | 0.033 | PVX_088015 | S | 163 | 0.502 | 0.027 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_088015 | T | 172 | 0.671 | 0.674 | PVX_088015 | S | 170 | 0.571 | 0.178 | PVX_088015 | T | 8 | 0.566 | 0.219 | PVX_088015 | S | 181 | 0.547 | 0.269 | PVX_088015 | S | 167 | 0.539 | 0.230 | PVX_088015 | S | 179 | 0.535 | 0.033 | PVX_088015 | S | 163 | 0.502 | 0.027 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_088015 | 328 S | LFVHSRHDG | 0.994 | unsp | PVX_088015 | 328 S | LFVHSRHDG | 0.994 | unsp | PVX_088015 | 328 S | LFVHSRHDG | 0.994 | unsp | PVX_088015 | 222 S | SKTHSEKLN | 0.993 | unsp | PVX_088015 | 247 S | ASRASYKFL | 0.994 | unsp |