

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
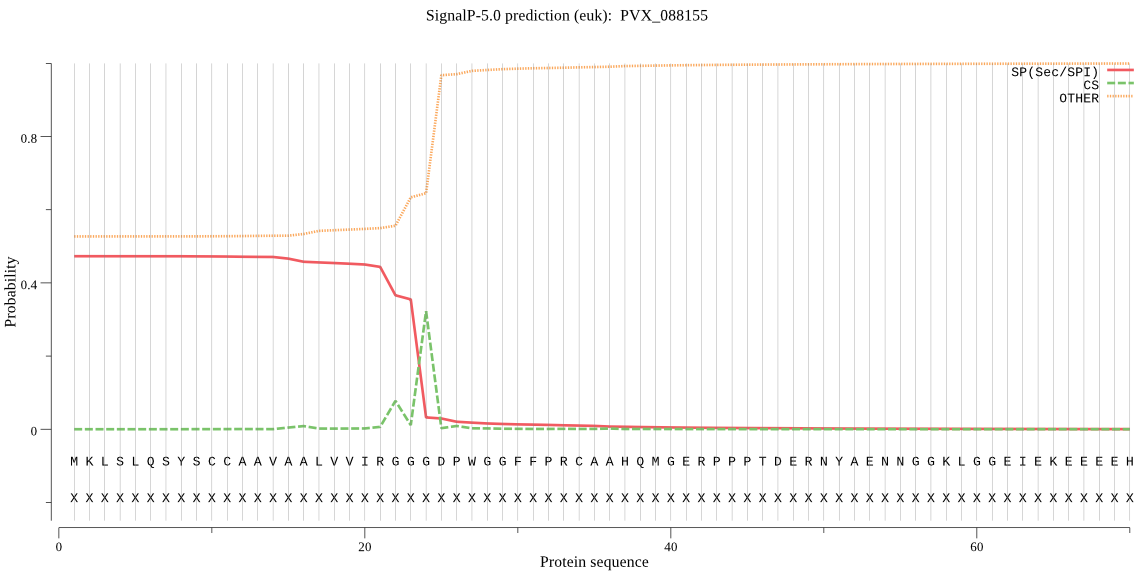
| PVX_088155 | SP | 0.253209 | 0.723081 | 0.023709 | CS pos: 22-23. IRG-GG. Pr: 0.4962 |
MKLSLQSYSCCAAVAALVVIRGGGDPWGGFFPRCAAHQMGERPPPTDERNYAENNGGKLG GEIEKEEEEHLHIDREVKGDDSHFTFDESSPTGGGKNVKVGLPLAKYTPGYVRPVLSDKR VTKGEAPGGENNLPSGDPQDGPPYLSKQQCGASFRDNAKADEHPSEGSHHDGEEPPSAAP NIWRRKKKNILQHRMSPTERGQEGYPPVEEKQTHREASNSDGSNLHESNFRGGSSYTEGS HQVGQLKLRTNRDDQKMMKGIFTPQVVRAMKEYLLKGGKKYGSGQVGHLGGSSDQQHGGD RRTGSNQAIHPLSGEAIHPLSSEAIHPLRFFPANFAPHSGDSQRGAARRSIINEKLRRNL SQITLSVSAAEERTKERVEEGAEVGAQSVAQSGGADDGEAGALRKIYKGVVKLYVDITEP SLETIWSNSPPKRVSGSGFVIEGGLILTNAHNVAYSTRILVRKHGCSKKYEGAVLHVAHE ADMALLTVADGSFYEDVSALELGPLPSLRDDVITVGYPSGGDKLSVTKGIVSRIEVQYYR HSNSRLLLTQIDAPMNPGNSGGPALVKGKVAGICFQLLKMANNTSYIIPTPVIKHFLMDL HRSGKYNGYPSLGVKYLPLDNANLRRLLGLTDLERRREVEENSGILVTEVDEEQMGCQSG GGGSTKWAVSSGEASTGAASSGEASMGAASTGAASSGEASMGAASTGAASHAITAGRGPL PGRPPITPTAPAEPLCYGLKKNDVLLSVEGTQIKSDGTVTLRGDESVDFQFLFNGKFVGD LCTCRVVRGGRVQTALVRVSRVHYLVRQHNWDVPNKFFIYGGVVFTTLTRSLYADEETDN VEVMRLLQFNLFKKQRDDEIVIVKRILPSKLTIGFNYQDCIVLTVNGIPVRNLQHLVGVI DGREGPLGGGVAKKSTQMKQMDQMGERRGTPSEGDANPYLKFTLEEDPFFQMKEEKDNLL HFLLLTSNGQQVPLVLLRAEVQRESAEIRRVYGITRERYVYSG
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| PVX_088155 | T | 691 | 0.525 | 0.211 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| PVX_088155 | T | 691 | 0.525 | 0.211 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_088155 | 218 S | HREASNSDG | 0.997 | unsp | PVX_088155 | 218 S | HREASNSDG | 0.997 | unsp | PVX_088155 | 218 S | HREASNSDG | 0.997 | unsp | PVX_088155 | 235 S | RGGSSYTEG | 0.991 | unsp | PVX_088155 | 435 S | PKRVSGSGF | 0.992 | unsp | PVX_088155 | 519 S | VGYPSGGDK | 0.994 | unsp | PVX_088155 | 670 S | KWAVSSGEA | 0.994 | unsp | PVX_088155 | 680 S | TGAASSGEA | 0.992 | unsp | PVX_088155 | 695 S | TGAASSGEA | 0.992 | unsp | PVX_088155 | 930 T | ERRGTPSEG | 0.996 | unsp | PVX_088155 | 932 S | RGTPSEGDA | 0.991 | unsp | PVX_088155 | 196 S | QHRMSPTER | 0.997 | unsp | PVX_088155 | 213 T | EEKQTHREA | 0.992 | unsp |