

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_088170 | OTHER | 0.999461 | 0.000272 | 0.000267 |
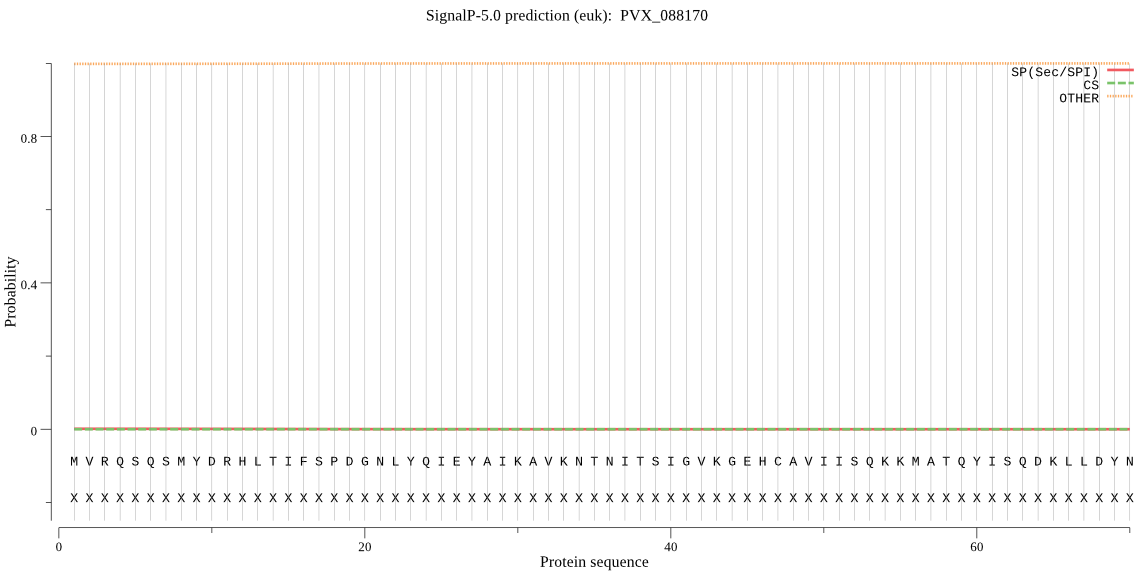
MVRQSQSMYDRHLTIFSPDGNLYQIEYAIKAVKNTNITSIGVKGEHCAVIISQKKMATQY ISQDKLLDYNNITNIYNITDEIGCSMVGMPGDCLSMVYKARIEAAEFLYSNGHNVNVETL CRNICDKIQIFTQHAYMRLHACSGMIIGMDEDNKPGLFKFDPSGFCAGYRACVVGNKEQE SISILERLLEKRKKKIQQETLEEDVQNTIILAIEALQTILAFDLKANEIEMAIVSKTNPN FTQISEKEIDNYLTFIAERD