

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_089485 | SP | 0.461768 | 0.535998 | 0.002234 | CS pos: 23-24. LNG-YI. Pr: 0.8471 |
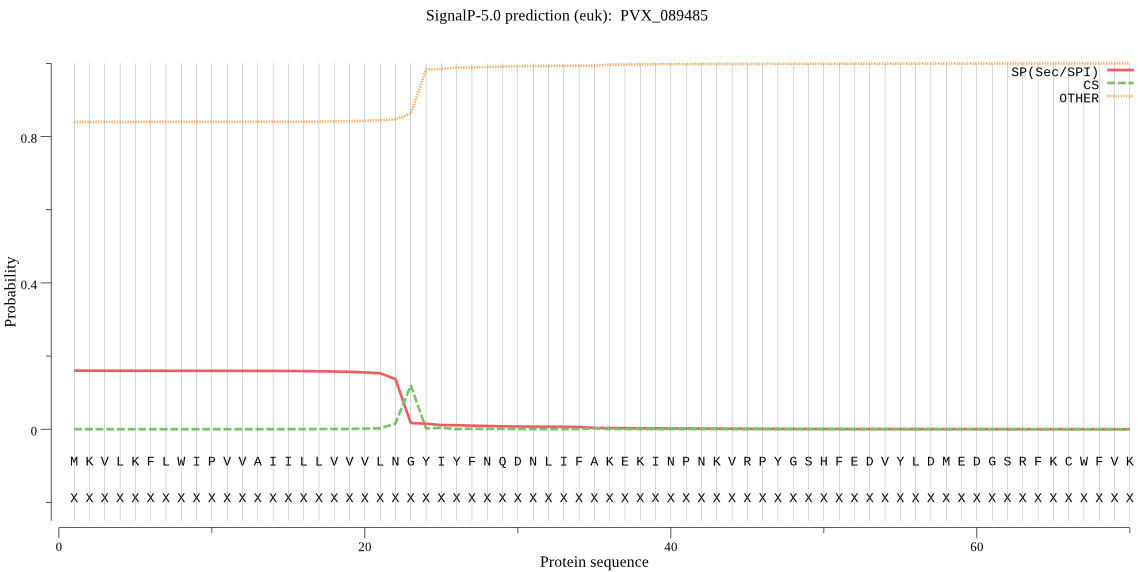
MKVLKFLWIPVVAIILLVVVLNGYIYFNQDNLIFAKEKINPNKVRPYGSHFEDVYLDMED GSRFKCWFVKAEDHENKPVFLYYLGKGGYVEKYVKLFDMIVQRVDVSIFSCSNRGCGTNK GNPSEEQLYKDALVPLNYLKQKNTKQLFIFGNSMGGAVALETASKHQKDLYGLILENPFL SVKDMSEQNYPFLNFFLLSFDVLIRTKMDNKEKIKKIHVPLLINTSELDKMIPPDHSSKL FELCPSKHKFRYFSKFGTHNNIIKVDDGSYHASLKKFVQTAISLREGKGEPQIPPPPRAA A
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PVX_089485 | 181 S | NPFLSVKDM | 0.996 | unsp | PVX_089485 | 283 S | QTAISLREG | 0.994 | unsp |