

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
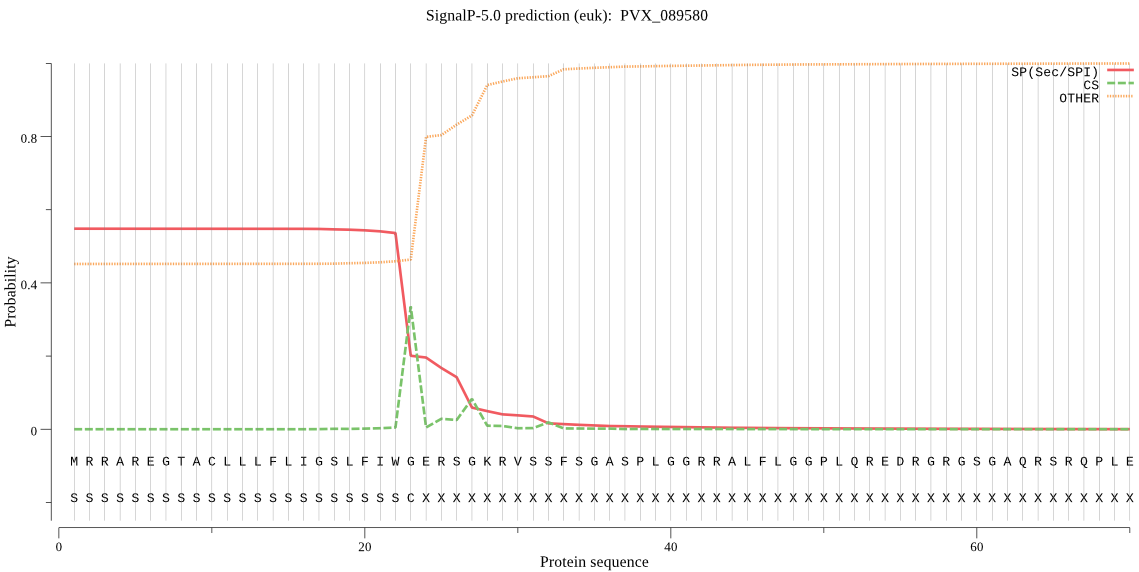
| PVX_089580 | SP | 0.135146 | 0.862394 | 0.002460 | CS pos: 23-24. IWG-ER. Pr: 0.6501 |
MRRAREGTACLLLFLIGSLFIWGERSGKRVSSFSGASPLGGRRALFLGGPLQREDRGRGS GAQRSRQPLEGAPKARFQEERAVGSRRGAASGRLFYSVGGPPGGGNSERQSVHPDERRGV HPPPRRGDRLAARVAPLRMSDEEYTINSDDYTEKAWEAISTLNKIGEKYESAYVEAEMLL LALLKDGPEGLAQRILKESGIDTELLIQEVDEYLKKQPKMPSGFGEQKILGRTLQAVLGT SKRLKKEFHDEYISIEHLLLGIVAEDSKFTRPWLLKYNVNYEKVKKATERVRGKKKVTSK TPEMTYQALEKYSRDLTALARAGKLDPVIGRDTEIRRAIQILSRRTKNNPILLGDPGVGK TAIVEGLAIKIVQGDVPDSLKGRKLVSLDLSSLIAGAKYRGDFEERLKSILKEVQDAEGQ VVMFIDEIHTVVGAGAVAEGALDAGNILKPMLARGELRCIGATTVSEYRQFIEKDKALER RFQQILVEQPSVDETISILRGLKERYEVHHGVRILDSALIQAAVLSDRYISYRFLPDKAI DLIDEAASNLKIQLSSKPIQLDHIEKQLVQLEMEKISILGDSPLGGGAKHTNPLNTPLNA APPEEPPVDYSQSPNFLKKKINEKEIARLKMIDHLMSQLRKEQRSILESWSSEKSYVDNI RAVKERIDVVKIEIEKAERYFDLNRAAELRFETLPDLEKQLKKAEDNYLNDIPERNRMLK DEVTSEDIMHIISLSTGIRLNKLQKSEKEKILNLDKELHKKIVGQDDAVRIVSKAVQRSR VGMNDPKRPIASLMFLGPTGVGKTELSKVLADVLFDTPDAVIHFDMSEYMEKHSISKLIG AAPGYVGYEQGGLLTDAVRKKPYSIILFDEIEKAHPDVYNLLLRVIEEGKLSDTKGNLAN FRNTIIIFTSNLGSQSILDLANDPNKKEKIKQQVMKSVRETFRPEFYNRIDDHVIFDSLS KKELKQIANIEIEKVANRLIDKNFKISIDDAVFSYIVDKAYDPAFGARPLKRVIQSEIET EIAVRILNETFVENDTIRVTLRDGALHFAKG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_089580 | 379 S | DVPDSLKGR | 0.991 | unsp | PVX_089580 | 379 S | DVPDSLKGR | 0.991 | unsp | PVX_089580 | 379 S | DVPDSLKGR | 0.991 | unsp | PVX_089580 | 531 S | DRYISYRFL | 0.993 | unsp | PVX_089580 | 746 S | KLQKSEKEK | 0.996 | unsp | PVX_089580 | 937 S | QVMKSVRET | 0.996 | unsp | PVX_089580 | 960 S | FDSLSKKEL | 0.997 | unsp | PVX_089580 | 987 S | NFKISIDDA | 0.993 | unsp | PVX_089580 | 1016 S | RVIQSEIET | 0.991 | unsp | PVX_089580 | 140 S | PLRMSDEEY | 0.997 | unsp | PVX_089580 | 199 S | ILKESGIDT | 0.997 | unsp |