

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_091410 | OTHER | 0.999835 | 0.000103 | 0.000062 |
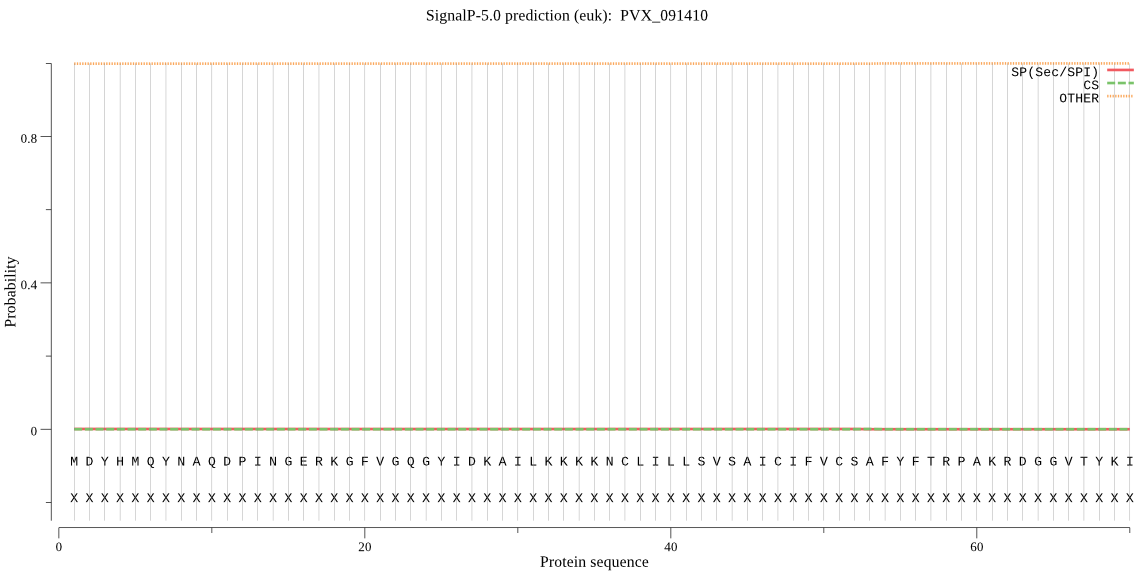
MDYHMQYNAQDPINGERKGFVGQGYIDKAILKKKKNCLILLSVSAICIFVCSAFYFTRPA KRDGGVTYKIADKVDDDYIINFLLKSKNGKKFIASKVQELISTYDVKKENLPAQHGGTYK RFAKRDQCTRNNCSVSPYGKNAAQVEVVAQVNLVNPLVDTKFLMANLETVNSFYLFMKEH GKEYSTADEMQQRYLSFAENLAKIKAHNSRENVLYRKGMNRFGDLSFEEIKKKYLTLKSF DLKSDGIKSPRVSDYDDIIHKYKPKDGTFDYVKHDWREFNAVTPVKDQKNCGACWAFSTV GVVESQYAIRKKELVSLSEQEMVDCSFKNYGCDGGNIPIAFEDLLDLGGICKEKEYPYVD VTPELCDIDRCKNKYKITTYVEIPQLRFKEAIKFLGPISVSICANDDFVYYEGGLFDGSC GFSPNHAVILVGYGMEEMYDAMSRKNEKRYYFWLKNSWGEKWGEKGYMKIQTDEYGLMKT CSLGAQAFVALIDEV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_091410 | 253 S | SPRVSDYDD | 0.998 | unsp | PVX_091410 | 253 S | SPRVSDYDD | 0.998 | unsp | PVX_091410 | 253 S | SPRVSDYDD | 0.998 | unsp | PVX_091410 | 316 S | KELVSLSEQ | 0.996 | unsp | PVX_091410 | 443 S | YDAMSRKNE | 0.997 | unsp | PVX_091410 | 226 S | FGDLSFEEI | 0.994 | unsp | PVX_091410 | 249 S | DGIKSPRVS | 0.993 | unsp |