-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs: GO:0003824
-
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>PVX_091434
MSRKRVFLLCIFLAISAVVDEAESFRKPDAAANVSFLEFTADGNAEEGQAQPSEPIVNTQ
PLQENVSEAAPHSEESTGGSAEKGSGGHEETPHAGEHDKGSGGSNGSSESNGSVLADGHA
EEAPNEQVSSEKNDGHSESSTTGSHSDKEELDLYTSSPSDKEENSAPAMGGMHNTVGLRG
GSASESAAPSEEKASEENVPHEVADKNGEDTSQEGSHAGALSPAHVGGESGESGGEHGEH
GEHAEHGEHGEHGEHGEHAEHGEHGEHGEHAEHGEHGEHGEHGEHGEHAEQEEHQIYGTI
HPEAEASILPALKEKGIHVSSPERVILEIIESAKEGIKGLLKLRKSKDTGKLLEEALEKL
NINQRDLHANNNFITLEMYDRILSEMFKILTEMSFYKDGHFYETLGLNKSILNQSLKEIK
IKMLRTIGVPYTKLPPIVKNKEKESTCAANNLIISITSKELAQRMAIMFAKWLAPEEYGS
VVDLDKSIELNVLCAGAPILVQQWKYYQNMLGFETGNEHAFLNLIDELLVIDKRHSNNEA
YSKVIRKIKKSKAFNYCTKVMRIAGNISSIPFNHENNKTPSYSIIGSLGNLVKAHMGNYY
VAIANRINSYFAYAEKRNKKNSPLKVVSVCTLLHLTDMLHNCSDEHLKNILDLNTLKLNI
LNMQGRRVLQPLVKMSFLGAAQNPALKEICEPSNHLVDETLSKLLNLLSTGSHELLAAEV
EKRGFDEDYIQEEIKNINESDNNVRDKGEDEVENLIFEDL
- Download Fasta
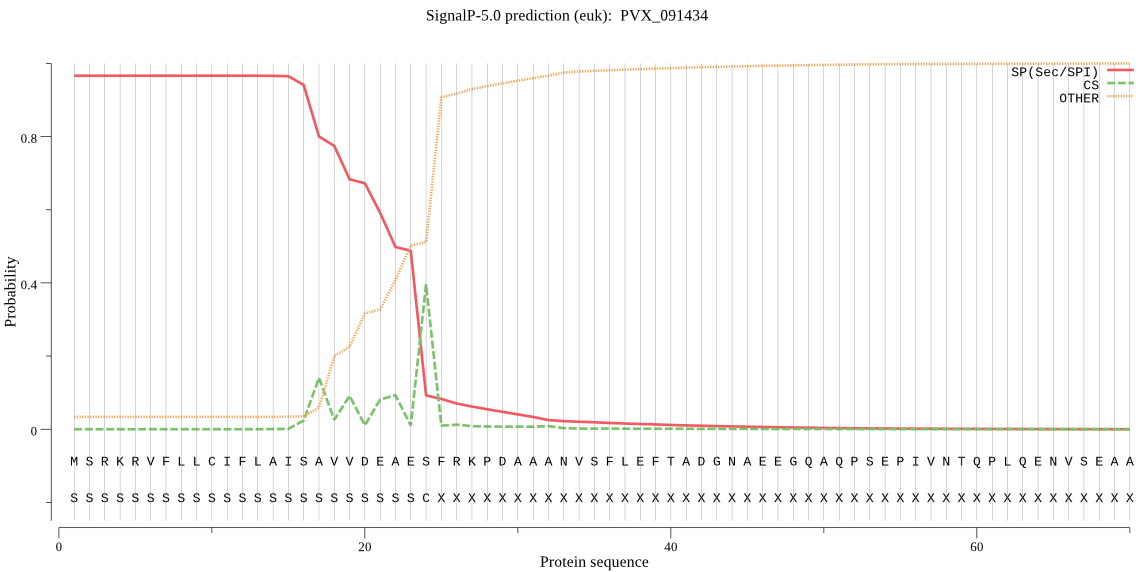
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/PVX_091434.fa
Sequence name : PVX_091434
Sequence length : 760
VALUES OF COMPUTED PARAMETERS
Coef20 : 5.108
CoefTot : 1.029
ChDiff : -54
ZoneTo : 19
KR : 3
DE : 0
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.165 2.276 0.269 0.714
MesoH : -0.639 0.314 -0.424 0.251
MuHd_075 : 20.035 5.642 4.738 3.478
MuHd_095 : 6.399 8.242 4.224 2.274
MuHd_100 : 0.993 4.665 2.407 1.814
MuHd_105 : 4.363 5.955 1.039 2.480
Hmax_075 : 16.887 15.750 2.575 5.160
Hmax_095 : 14.525 18.637 4.295 5.031
Hmax_100 : 13.000 17.800 3.802 5.800
Hmax_105 : 15.283 17.150 1.685 5.215
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.9601 0.0399
DFMC : 0.9538 0.0462
-
Fasta :-
>PVX_091434
ATGTCTCGTAAAAGGGTTTTTCTTCTGTGCATTTTTCTGGCCATTTCCGCCGTTGTTGAC
GAAGCCGAGTCGTTCAGGAAGCCAGACGCGGCAGCGAACGTCTCCTTCCTGGAGTTCACA
GCAGATGGCAACGCTGAAGAGGGGCAGGCGCAGCCAAGTGAACCCATCGTTAACACCCAG
CCGCTGCAGGAAAATGTAAGTGAAGCCGCTCCCCACTCGGAGGAGTCCACTGGGGGAAGC
GCGGAGAAGGGAAGTGGCGGCCACGAGGAAACCCCCCACGCAGGAGAACATGACAAAGGT
AGTGGCGGAAGCAACGGGAGCAGCGAAAGCAACGGAAGCGTCTTGGCTGATGGCCACGCC
GAAGAAGCGCCGAACGAGCAGGTGAGTAGCGAGAAGAACGACGGACACAGTGAAAGCAGC
ACCACGGGCTCACACTCAGATAAGGAGGAGCTGGACTTGTATACGTCCTCCCCCTCCGAT
AAAGAAGAAAACAGCGCCCCTGCCATGGGTGGTATGCACAACACGGTAGGCCTGAGGGGG
GGAAGCGCCTCTGAATCGGCGGCCCCAAGTGAAGAGAAAGCATCCGAAGAGAATGTCCCC
CATGAAGTGGCAGATAAAAATGGGGAGGACACCTCACAGGAGGGTTCCCACGCGGGAGCG
CTGTCACCTGCGCATGTGGGTGGCGAGAGTGGCGAGAGTGGCGGCGAGCACGGAGAGCAC
GGTGAACACGCTGAACATGGGGAGCACGGAGAGCACGGAGAGCACGGTGAACACGCTGAA
CATGGGGAGCACGGAGAGCACGGTGAACACGCTGAACATGGGGAGCACGGAGAGCACGGT
GAACATGGCGAGCACGGTGAACATGCTGAACAGGAGGAGCACCAAATCTATGGGACGATT
CACCCCGAGGCGGAAGCCTCCATTTTGCCCGCGCTGAAGGAAAAGGGCATCCACGTGTCC
AGCCCAGAAAGGGTAATCCTCGAGATCATCGAAAGCGCCAAGGAGGGAATAAAAGGATTG
CTGAAGCTCCGGAAGAGCAAAGACACAGGCAAGCTGCTGGAGGAGGCACTAGAAAAGCTA
AACATAAACCAAAGGGATCTGCATGCAAACAATAATTTCATTACCCTAGAAATGTATGAC
CGGATCCTCTCCGAAATGTTTAAAATATTAACAGAAATGTCATTCTATAAGGATGGCCAT
TTTTACGAAACGTTAGGTTTGAATAAATCTATACTAAATCAATCGCTGAAAGAAATTAAA
ATAAAAATGTTGAGGACCATTGGAGTGCCATACACGAAGCTACCGCCAATCGTTAAGAAC
AAAGAGAAGGAAAGCACATGTGCTGCGAACAATCTCATCATTAGCATTACATCAAAAGAG
CTAGCTCAACGAATGGCTATCATGTTCGCAAAATGGCTAGCTCCAGAAGAATACGGGTCT
GTCGTCGATTTAGATAAAAGCATAGAATTAAATGTCCTCTGCGCAGGGGCACCTATCCTA
GTACAGCAATGGAAATATTACCAAAACATGTTAGGGTTCGAAACGGGAAATGAACATGCC
TTCTTAAACCTCATCGATGAGTTGCTAGTCATCGACAAGAGACACAGTAACAATGAAGCG
TATTCAAAAGTGATAAGGAAAATAAAAAAATCGAAAGCCTTTAATTATTGTACTAAAGTT
ATGAGGATTGCAGGGAATATTTCTTCCATTCCATTTAATCATGAAAATAATAAAACGCCT
AGCTACTCCATCATCGGGTCGTTAGGTAACCTGGTGAAGGCTCACATGGGGAACTACTAC
GTTGCCATTGCTAATAGGATTAACTCCTATTTTGCCTATGCCGAGAAGAGGAACAAAAAA
AACAGCCCCTTAAAAGTTGTCTCCGTCTGTACTCTCCTTCATTTGACCGACATGCTCCAC
AATTGCTCTGATGAACATTTAAAGAATATTTTGGACCTCAACACTTTGAAGCTTAACATT
CTGAACATGCAGGGCAGGCGCGTGCTGCAGCCCCTCGTCAAGATGAGCTTCCTCGGCGCC
GCGCAGAACCCCGCCCTCAAGGAGATCTGCGAGCCGAGCAACCACCTAGTGGACGAGACC
CTCTCCAAGCTGCTCAACCTGCTCTCGACCGGTTCGCACGAACTCCTCGCGGCCGAGGTG
GAAAAGCGCGGCTTCGACGAGGACTACATCCAGGAGGAAATCAAAAACATAAACGAAAGC
GACAACAACGTTCGGGACAAGGGCGAAGACGAAGTGGAAAATCTCATTTTTGAAGACTTG
TGA
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
PVX_091434 | 137 S | NDGHSESST | 0.994 | unsp | PVX_091434 | 137 S | NDGHSESST | 0.994 | unsp | PVX_091434 | 137 S | NDGHSESST | 0.994 | unsp | PVX_091434 | 144 S | STTGSHSDK | 0.994 | unsp | PVX_091434 | 146 S | TGSHSDKEE | 0.998 | unsp | PVX_091434 | 159 S | TSSPSDKEE | 0.995 | unsp | PVX_091434 | 182 S | LRGGSASES | 0.993 | unsp | PVX_091434 | 195 S | EEKASEENV | 0.991 | unsp | PVX_091434 | 233 S | ESGESGGEH | 0.994 | unsp | PVX_091434 | 415 S | ILNQSLKEI | 0.991 | unsp | PVX_091434 | 480 S | EEYGSVVDL | 0.993 | unsp | PVX_091434 | 536 S | DKRHSNNEA | 0.997 | unsp | PVX_091434 | 73 S | AAPHSEEST | 0.993 | unsp | PVX_091434 | 129 S | NEQVSSEKN | 0.994 | unsp |


