

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
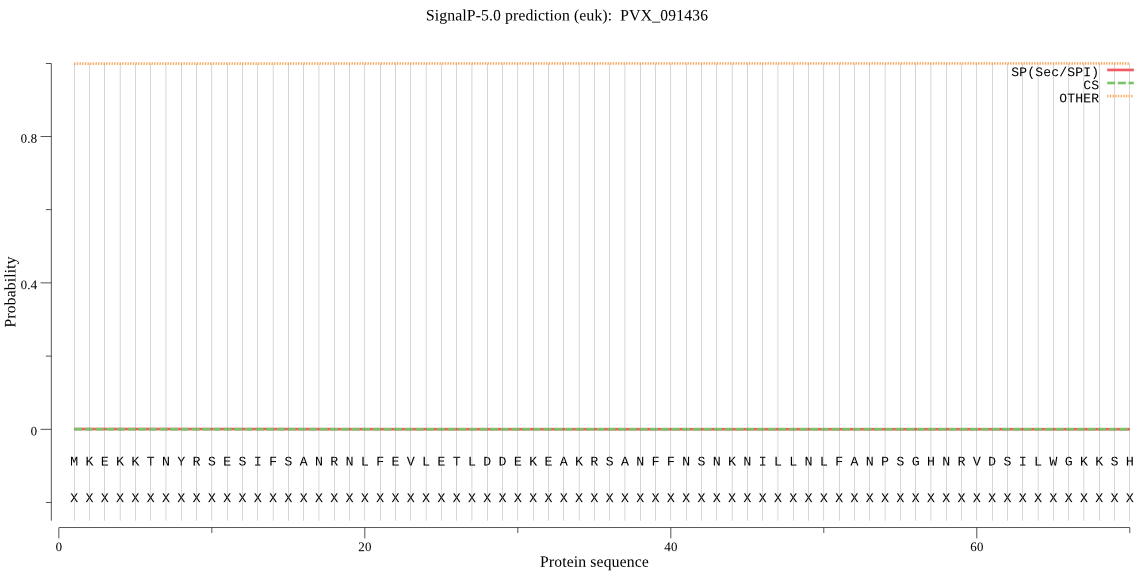
| PVX_091436 | OTHER | 0.999966 | 0.000008 | 0.000026 |
MKEKKTNYRSESIFSANRNLFEVLETLDDEKEAKRSANFFNSNKNILLNLFANPSGHNRV DSILWGKKSHLHHGRNARRKGCKKGEPRKGEKGKEETISGETIKGETLNDGPINDEPIND EPINGEPINEDAKTVRDSPICGSKKWIEHFCWFCSLNLLDEVLEKEIYKEKIHPHVYDLI YDENTYFDTSSMMSVPYEFARFMLTQLDNNVLHDIKINNIIKEEKYLFKKFCRKYKCDKK YRKVRGGPRSGRPPFSRGSSGGRGSNGSSGSSGRISSDSVKSNHSLLYRSRTVSVRYSNY LNIRRELFNSGRVGGKRVGGSSLPARKLFYESGNLGNCTGTDLFYCRRAAPPGEVSSGDG NISGGNRSGNRSNNKSCDTLSLDGEGKGRREAGKRSRLAAGGDAPNRRGGEQVSPSVSNR SGSGYSVGRVPPGERRGKLERSLTSRREMCRQRFAKTDVRNGVHVKGDKQCSRVLRRSNS FSMHHREGGIAANGGMANGGGPPQEARKDRPSSRSQRIYHKYKHNLNYIKQFYYVDETQK KPFPLCLYNIFKAHGTIKQCSRCYCFFNSTSCHCLECKRKSNLSLKNPHYFIFQHGLTAS VQDFQNIVNPLLIKYPHLFIYITYSNQSHTFEGVDVGTERICTELNCLFKIINYKINVSM IGHSLGGILNRSVLINLYRKKMFKNKRLINFITFACPHIGVHENMAIMKLLSTYLGAHTI DDLNNKTTLLLKIASVESISILKKFENIIFYGNAQSDWLVGIRTSLILPYTLFNEELILF IIEQAKNVPEIPINIFSVVHLYMRKKKLLFFYFYQDLKNPNYLLNKRKDQNRFLDQMLQT IISSRKLLSCSQRKKFNAFVDYYGGVGAKEGQSKGKPKRKKKSPSGGEAKGGSPSPGDAQ EGSAFPGDAHPGDADSSNSPSACPKREDSPPCEEQTSGSAPTQMNRRGSSIHTDGEAPEM RSTRDKPYMSDGVNVQMLPIHFRKNVSTVETNRRARVGVSHSCDKLPPMNVLSSGGKEGI CAEEDTCAKEDGNASGKSLTCEDVHLGREEANEKASANEKEKANVSAKEQANRASYYDRY LCIPESYKIASSSLAAERGGGPHSGANESGNREGSHREGSHREGSHREGSHREGNRPASN SRKKKLGDYKHFEKLFFECLKRKIINDIKTLDNNLKRKKKKADVAPSKSFSLQNTINALK NYSLFYLKNGAERGAHEGGGLDGGGLQSGRHDYQPIADGTLGEALEATHSVERESCEGNT NKQSNETHTDESNRKKESSTLQLCDANSEEYDYISEFFYTTSEDNEGGDANDESLGDEPV LARGRKSNPGGEHPDEQQSEHLDDQPPHNPGKSAPKKRWKGTTLLKGITKNDKKKYKQIL SHIYTISNEELIEKFCKNPELLYYEVLFYCLNQLPIQRYTISLPLYSNAHVQIIAHPRIC SGESATVKHFVEHLIL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_091436 | 279 S | ISSDSVKSN | 0.997 | unsp | PVX_091436 | 279 S | ISSDSVKSN | 0.997 | unsp | PVX_091436 | 279 S | ISSDSVKSN | 0.997 | unsp | PVX_091436 | 445 S | RSLTSRREM | 0.995 | unsp | PVX_091436 | 512 S | KDRPSSRSQ | 0.998 | unsp | PVX_091436 | 513 S | DRPSSRSQR | 0.99 | unsp | PVX_091436 | 584 S | KSNLSLKNP | 0.995 | unsp | PVX_091436 | 883 S | RKKKSPSGG | 0.998 | unsp | PVX_091436 | 885 S | KKSPSGGEA | 0.997 | unsp | PVX_091436 | 895 S | GGSPSPGDA | 0.996 | unsp | PVX_091436 | 929 S | KREDSPPCE | 0.996 | unsp | PVX_091436 | 949 S | NRRGSSIHT | 0.997 | unsp | PVX_091436 | 962 S | PEMRSTRDK | 0.995 | unsp | PVX_091436 | 1066 S | KANVSAKEQ | 0.996 | unsp | PVX_091436 | 1115 S | NREGSHREG | 0.998 | unsp | PVX_091436 | 1120 S | HREGSHREG | 0.997 | unsp | PVX_091436 | 1125 S | HREGSHREG | 0.997 | unsp | PVX_091436 | 1130 S | HREGSHREG | 0.996 | unsp | PVX_091436 | 1139 S | NRPASNSRK | 0.994 | unsp | PVX_091436 | 1255 S | VERESCEGN | 0.996 | unsp | PVX_091436 | 138 S | TVRDSPICG | 0.998 | unsp | PVX_091436 | 260 S | SRGSSGGRG | 0.995 | unsp |