

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
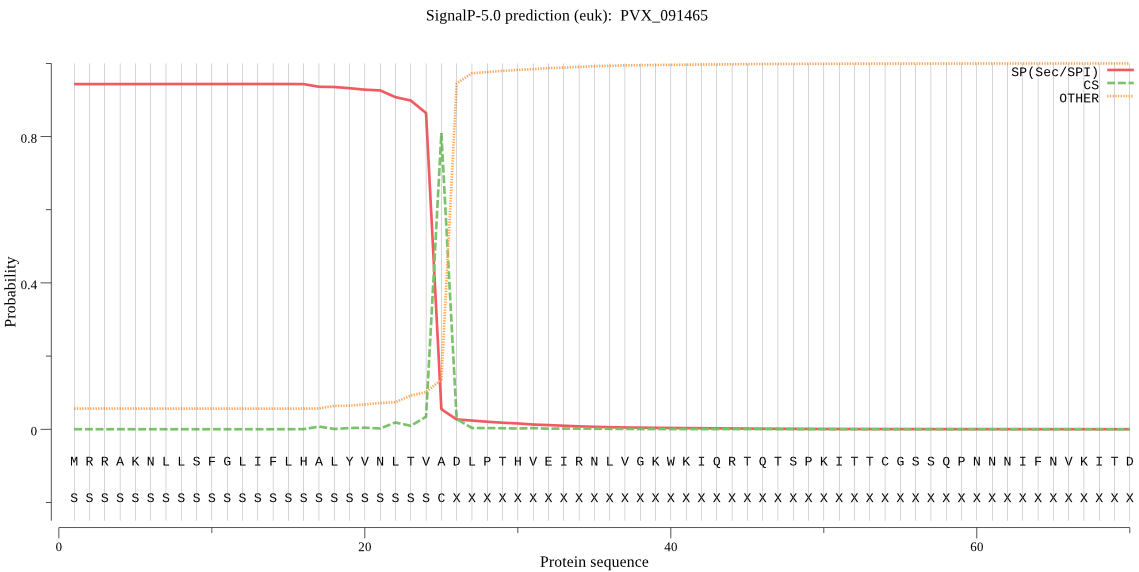
| PVX_091465 | SP | 0.002900 | 0.997004 | 0.000096 | CS pos: 25-26. TVA-DL. Pr: 0.9559 |
MRRAKNLLSFGLIFLHALYVNLTVADLPTHVEIRNLVGKWKIQRTQTSPKITTCGSSQPN NNIFNVKITDYKKYLLDNHYKFTSELYVILSDDFVLYGDMHDTTGNEHRKNWKVLVVYDE HKRKIGTWTTICDEGFEIRLGNETYTALMHYEPTGKCGEAKEDDATDSNGETECYVTNYN KIRFGWVDISNTKDEKLYGCFYAERYRVSDEPASAVEVSHNFAKETTANSSVLKVHNFLY PPPSASTSSSSDKPTFTKRKNMQIDQNSELYWHKMKHHGKKKPITKAMIMNAKQRYACPC NKDEHVQNEVNDGDNPDQPVSPVSPVSLMQLGAGSGEDGTNEMDLENYEDTEKAPHRELE IDELPKNFTWGDPFNNNTREYDVTNQLLCGSCYIASQMYVFKRRIEIGLTKNLDKKYLNN FDDLLSIQTVLSCSFYDQGCNGGYPYLVSKMAKLQGIPLDKVFPYTATAETCPYPVDQAG PMTTPAPGAANRGSGSAKPLNNLRQINAVIFGKRTTNDMHEHFESPISDDPDKWYAKDYN YIGGCYGCNQCNGEKIMMNEIFRNGPIVASFEASPDFYEYADGVYYVEDFPHARRCTVDD TKSNFVYNITGWEKVNHAIVLVGWGEEEIDGALYKFWIGRNSWGKTWGKEGYFKIIRGVN FSGIESQSLFIEPDFTRGAGKILLEKLRNE
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| PVX_091465 | T | 484 | 0.570 | 0.661 | PVX_091465 | T | 483 | 0.558 | 0.185 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| PVX_091465 | T | 484 | 0.570 | 0.661 | PVX_091465 | T | 483 | 0.558 | 0.185 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_091465 | 251 S | TSSSSDKPT | 0.997 | unsp | PVX_091465 | 251 S | TSSSSDKPT | 0.997 | unsp | PVX_091465 | 251 S | TSSSSDKPT | 0.997 | unsp | PVX_091465 | 335 S | LGAGSGEDG | 0.995 | unsp | PVX_091465 | 168 S | DATDSNGET | 0.991 | unsp | PVX_091465 | 249 S | ASTSSSSDK | 0.993 | unsp |