

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
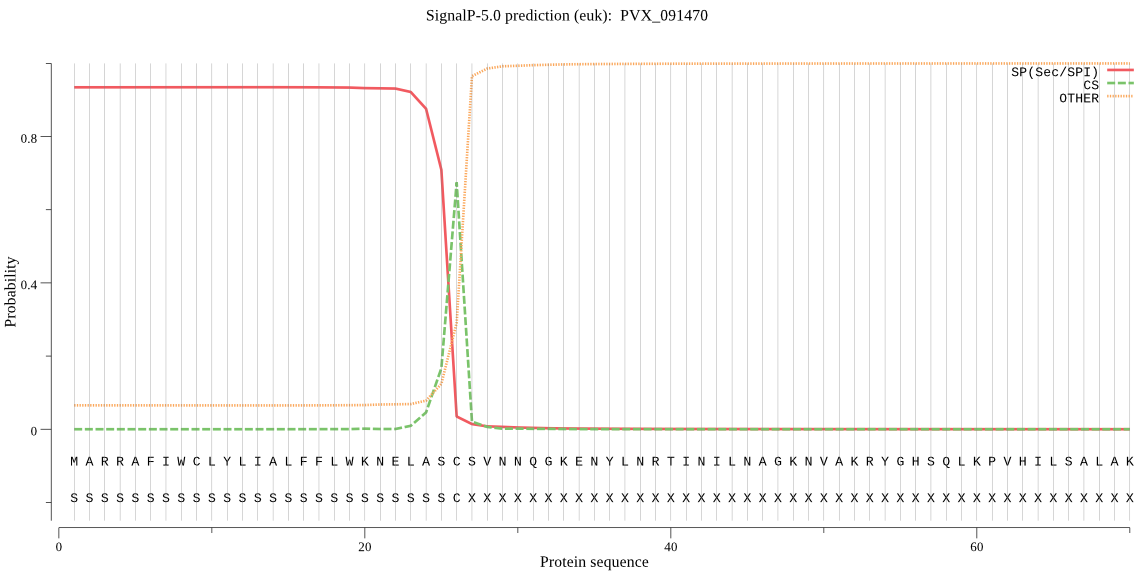
| PVX_091470 | SP | 0.014141 | 0.985499 | 0.000360 | CS pos: 26-27. ASC-SV. Pr: 0.9010 |
MARRAFIWCLYLIALFFLWKNELASCSVNNQGKENYLNRTINILNAGKNVAKRYGHSQLK PVHILSALAKSDYGSNLLKENSVNASNLKEYIDTALEQTRAGAPLDNKSKIAYSDEVKEV LAEAEALASKYKSQKVDVEHLLSGLMNDELVNEIMNEVYLTEEAVKGIMKSKFEKTKKDK DGKSGGLYLEQFGSNLNEKVRNGKLQGIYGRDEEIRAVIESLLRYNKNSPVLVGQPGTGK TTIVEGLVYRIEKGDVPKELRGYTVISLNFRKFTSGTSYRGEFETRMKNIIKELKNKKNK IIIFVDEIHLLLGAGKAEGGTDAANLLKPVLSKGEIKLIGATTIAEYRKFIESCSAFERR FEKILVEPPSVENTIKILRSLKSKYENFYGIHITDKALVAAAKVSDRFIKDRFLPDKAID LLNKACSFLQVQLSGKPRIIDVTEREIERLAYEISTLEMDVDKVSKRKYNNLIKEFENKK ELLKKYYEEYVISGERLKRKKETEKRLNELKELAQNYIIANKEPPIELQNSLKEAQEKYM DVYKETLAYVEAKTHNAMNVDAVYQEHVSYIYLRDSGMPLGSLSFESSKGALKLYNSLSK SIIGNEDIIKSLSDAVVKAATGMKDPEKPIGTFLFLGPTGVGKTELAKTLAIELFSSKDN LIRVNMSEFTEAHSVSKITGSPPGYVGFSDSGQLTEAVRERPHSVVLFDELEKAHPDVFK VLLQILGDGYINDNHRRNIDFSNTIIIMTSNLGAELFKKKLFFDANNSDTPEYKRVFDDL RIQLIKKCKKVFKPEFVNRIDKIGVFEPLSKKNLREIVKLRFKKLEKRLEEKNIHVSVSE KAVDYIIDQSYDPELGARPTLIFIESVIMTKFAIMYLKKELVDDMDVHVDFNKAANNLVI NLSTV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_091470 | 370 S | VEPPSVENT | 0.991 | unsp | PVX_091470 | 370 S | VEPPSVENT | 0.991 | unsp | PVX_091470 | 370 S | VEPPSVENT | 0.991 | unsp | PVX_091470 | 531 S | ELQNSLKEA | 0.995 | unsp | PVX_091470 | 656 S | IELFSSKDN | 0.996 | unsp | PVX_091470 | 704 S | ERPHSVVLF | 0.994 | unsp | PVX_091470 | 839 S | HVSVSEKAV | 0.991 | unsp | PVX_091470 | 133 S | SKYKSQKVD | 0.995 | unsp | PVX_091470 | 278 S | TSGTSYRGE | 0.992 | unsp |