

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
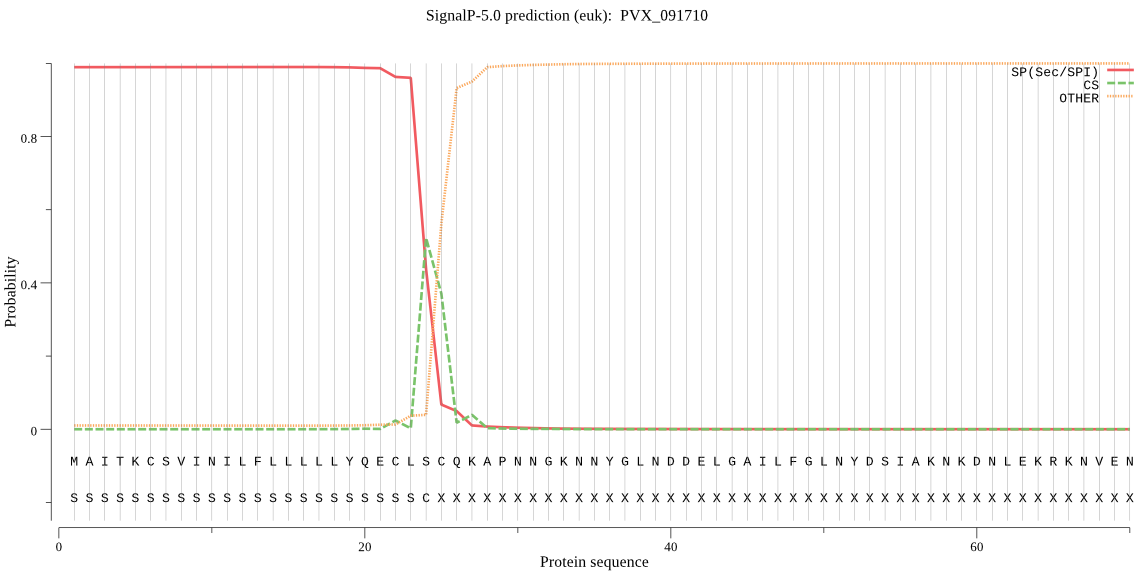
| PVX_091710 | SP | 0.039958 | 0.960005 | 0.000037 | CS pos: 24-25. CLS-CQ. Pr: 0.5303 |
MAITKCSVINILFLLLLLYQECLSCQKAPNNGKNNYGLNDDELGAILFGLNYDSIAKNKD NLEKRKNVENESIFLRNFANEDTSKNTQSEKAQKEIKIETETESVNSNEKEVATSQKSDT SNKNSSVENEKIELKNDELLGKNFEKDKVNKKGDNTNTTNNHDLTNSSEKQGVDIRGSKN MNNYLQKTGDTNIEKSESLQKDVNIKNHNEEANDAKRLDSAQTNNEKSKISKDTIDKDVQ SNELTNLASNRSNKKSQGLAKKENELKSANLEENHNAKKDLLKKDQKREDGKKITHPENS NSDQYGVQVSLNDEEKNTNTKSVSHSEDHSASYSGEKFGTHVSNSQKDMLKNIRPVQFDE SAYGKLNGGSPENDENEILNKINKNNENNFSEKVALRKGTKDRNEYEYFKLKSNDFKVLG IINKYSSRGGFSISVDCGGYDDFDEVPGVSNLLQHAIFYKSEKRNTTLLSELGKYSSEYN SCTSESSTSYYATAHSEDIYHLLNLFAENLFYPVFSEEHIQNEVKEINNKYISIENNLES CLKIASQYITNFKYSKFFVNGNYTTLCENVLKNRLSIKNILTEFHKKCYQPRNMSLTILL GNKVNTADHYNMKDVENMVVHIFGKIKNESYPIDGDVIGKRINRMESERVNLYGKKDSYN DANFIHIEGRNEKEAAFLQSMNELHYALDLNQKSRYVEIIKKEEWGDQLYLYWSSKTNAE LCKKIEEFGSMTFLREIFSDFRRNGLYYKISVENKYVYDLEVTSICNKYYLNFGILVKLT QRGRTNLAHLIHICNVFVNEIGKLFDRDSLDKGISKYILDYYREKALVTDLKFNSDNVNV SLDDLVIYSKRLLVHADDPSSLLTIHSLIEDKHKNDFRNHIKITSLVGSLIRNDNLNIIN VVDAFSLTNQSQIPNTAITYALGENPYLVGQGGVTNDVNIALPEIKVCPFNSFDHNDNNV LHEESFFCVPYNNRENFEFSDTKPMFVSEENKNEFRASVLYSMPCLIKSSYGYNVYFKRG LTDVSKVKADFIFYFPSEKFTFYEAIFSRIHIIILRKKIKLFLTDYASCSVNVHIKRNAE SYTVHVDTNSYYFAELFSKLGDLLSVKEIPSKEEFDDAYDELIMNVRTSRKLGAENSLRI MHSLFNKYEPTDKEMDDILSAYFFYPSYKDYTRYVVDIFHRNYVSIFIYGNIAMPTEKED ENATSHTDQSATNGGSGVPNSSSSHVNDNVKDNGNDNDNTNTKANVGEHRSSLTLYAEAN NTGSASPWEKRDHRLDAQGQLGEQAFAADSIQISHNGVGIEYLVNLSESFIKKVTNGVVR RSESTYYTSTLVHNEDVEINIPHMSQNGSSSIIVSYTIESESMLSDVLINIIVDLISSEF VKVSKLRYNDGYMVEVKPFFARYGLSGLLFVIQSFEKDVEKLEAHICTFVKYVIFQLMNI GTSDLVKKLESMKEYYIINNSIFTFDQEYASITDQLTSGMECFDKKYKIIKIFDELINCP KMIINKMNAILKNSKKAIFKEYKAAKAVDAPSSKSDEVDDANSYVCNYSRKAPAKLSSVQ LAANSRSAKKKQPKERTLRKDTLRMRKPHKMANFISVSNFVEIKRKGFFQYVIDYFRSNV NSPLNDSSYLDFKSCDEEMSKRNFRVFYNFANDVNKIREYFLLKFASDKEVREKCSVDYE EIRQYCSGH
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_091710 | 125 S | SNKNSSVEN | 0.993 | unsp | PVX_091710 | 125 S | SNKNSSVEN | 0.993 | unsp | PVX_091710 | 125 S | SNKNSSVEN | 0.993 | unsp | PVX_091710 | 126 S | NKNSSVENE | 0.997 | unsp | PVX_091710 | 300 S | HPENSNSDQ | 0.993 | unsp | PVX_091710 | 324 S | TKSVSHSED | 0.992 | unsp | PVX_091710 | 345 S | HVSNSQKDM | 0.998 | unsp | PVX_091710 | 370 S | LNGGSPEND | 0.993 | unsp | PVX_091710 | 461 S | IFYKSEKRN | 0.992 | unsp | PVX_091710 | 489 S | ESSTSYYAT | 0.994 | unsp | PVX_091710 | 576 S | KNRLSIKNI | 0.992 | unsp | PVX_091710 | 647 S | NRMESERVN | 0.994 | unsp | PVX_091710 | 1111 S | KEIPSKEEF | 0.994 | unsp | PVX_091710 | 1129 S | NVRTSRKLG | 0.997 | unsp | PVX_091710 | 1167 S | FFYPSYKDY | 0.992 | unsp | PVX_091710 | 1205 S | ENATSHTDQ | 0.996 | unsp | PVX_091710 | 1451 S | KKLESMKEY | 0.996 | unsp | PVX_091710 | 1471 S | QEYASITDQ | 0.995 | unsp | PVX_091710 | 1532 S | VDAPSSKSD | 0.994 | unsp | PVX_091710 | 1533 S | DAPSSKSDE | 0.995 | unsp | PVX_091710 | 1535 S | PSSKSDEVD | 0.993 | unsp | PVX_091710 | 115 S | EVATSQKSD | 0.996 | unsp | PVX_091710 | 121 S | KSDTSNKNS | 0.993 | unsp |