

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
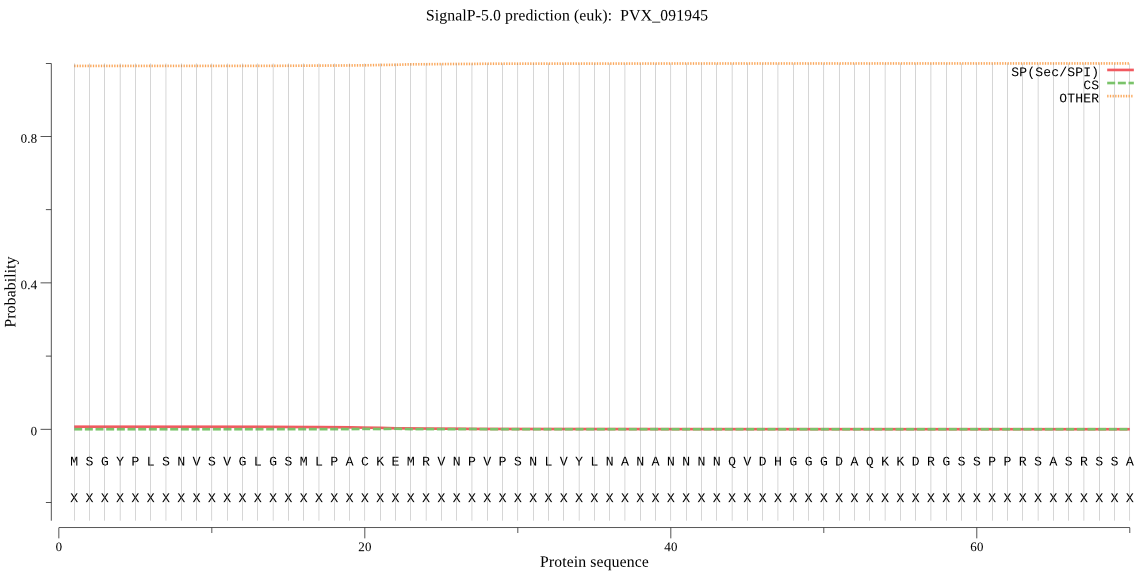
| PVX_091945 | OTHER | 0.993586 | 0.003831 | 0.002582 |
MSGYPLSNVSVGLGSMLPACKEMRVNPVPSNLVYLNANANNNNQVDHGGGDAQKKDRGSS PPRSASRSSANQSNSKNDAKDEKQALECTSYKIGKGKKKNNNLDSMEELLFKLTNGNFKA EKHHVYTADGYRLNLYRIVSTNKKDNLQKKKEVFCLNHGLFESSISYTCKGYESLAFQIF ANDYDVWISNNRGNAFTKYVGKDYALKKLKERYSLQDLKDIGVEVTEEMAAQGSSSASGD DEKKSTESASSSDEEGCREKGKNNNLRKAGKADDNAANNSSGKKNSEGTKSSVEESKSVP PCCGTPGAKPSIGDASPAEGGSKVGSNKAEKSSPCTSSSSDSEGPEDAPANIDSRALSDI FKRAKGSKGADKNESIKKVYCGIGIPDEVDPSLSADDESTVNSEGEKQEEDELDEGDYNL GYEIDGSPDPPGPFGDSSSCDSNMVCSFPYVPKDPENGEAAGEAAECAEGVEDGEEVEEL VNLNKHSEEDDVVELNVPEMDNWTFEDMGTKDLPAVIKYIKNKTQREQIVYVGFSQGSVQ LLIGCCLNDYLNNSIKRTYLLSLPIILKNKDKLLKSVKMLLIASRWYKAIIGSKEFIQKV FPEKMSTSMISSSADLFTRNFFKLYTENVDENYKKIYFRHTPSGTTSKANLKKWCSSLHD GPVSEAIDKYAHKCSFPITLVYGIKDCLVDAERSIEYMKKKFTKNDLKIISEPEWSHIDP VLADNRNVVLSCILEDLKGEEKETKQEEEKETKQEEKETKQEEEKETKQEEKKQTKQEEK KQTKQEEKKKQQEEKKKQLKK
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| PVX_091945 | T | 305 | 0.549 | 0.083 | PVX_091945 | T | 336 | 0.531 | 0.050 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| PVX_091945 | T | 305 | 0.549 | 0.083 | PVX_091945 | T | 336 | 0.531 | 0.050 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_091945 | 75 S | NQSNSKNDA | 0.993 | unsp | PVX_091945 | 75 S | NQSNSKNDA | 0.993 | unsp | PVX_091945 | 75 S | NQSNSKNDA | 0.993 | unsp | PVX_091945 | 214 S | KERYSLQDL | 0.994 | unsp | PVX_091945 | 238 S | SSSASGDDE | 0.997 | unsp | PVX_091945 | 245 S | DEKKSTESA | 0.996 | unsp | PVX_091945 | 248 S | KSTESASSS | 0.991 | unsp | PVX_091945 | 250 S | TESASSSDE | 0.997 | unsp | PVX_091945 | 251 S | ESASSSDEE | 0.996 | unsp | PVX_091945 | 252 S | SASSSDEEG | 0.997 | unsp | PVX_091945 | 292 S | GTKSSVEES | 0.997 | unsp | PVX_091945 | 316 S | IGDASPAEG | 0.99 | unsp | PVX_091945 | 338 S | PCTSSSSDS | 0.992 | unsp | PVX_091945 | 340 S | TSSSSDSEG | 0.994 | unsp | PVX_091945 | 342 S | SSSDSEGPE | 0.994 | unsp | PVX_091945 | 358 S | SRALSDIFK | 0.99 | unsp | PVX_091945 | 487 S | LNKHSEEDD | 0.996 | unsp | PVX_091945 | 554 S | YLNNSIKRT | 0.99 | unsp | PVX_091945 | 60 S | DRGSSPPRS | 0.997 | unsp | PVX_091945 | 64 S | SPPRSASRS | 0.991 | unsp |