

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_092055 | SP | 0.041186 | 0.955578 | 0.003235 | CS pos: 20-21. AVA-PT. Pr: 0.3180 |
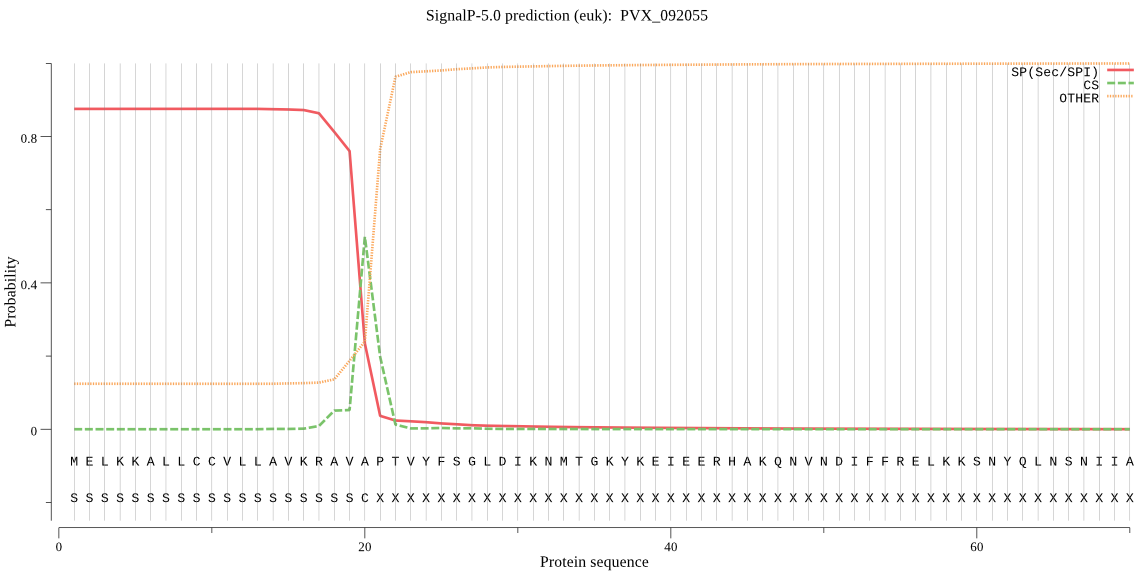
MELKKALLCCVLLAVKRAVAPTVYFSGLDIKNMTGKYKEIEERHAKQNVNDIFFRELKKS NYQLNSNIIALSTSRHYFNYRHTSNLLTAYKYLKHVGGNLDRNILLMVPFDQACNCRNIV EGTIFNEYEKPPSEDLKKKKMKENLYSHLHIDYKNDNIRDEQIRRVIRHRYDALTPVKNR LYTNGNRERNLFIYMTGHGGVSFFKIQDFNIVSSAEFSLYIQELLIKNIYKYIFVIIDTC QGYSFYDKTLHFLKKKKINNVFLMSSSDKNENSYSLHSSKYLGVSTVDRFTFYFFSYLEN MNRVYAHEPHKNAKAFSLYSILNYLKTQHLISTPTINSSKFSASMFMHSKNILFYDSSGF YVSKDAEGNALEGLGKHRYDASQRTDKTATRSTCLGDLITCGHVKNEIHTHMGNLYSRSS YYSNVEVFFKEESHFTDYYFTAERFAGGYLLPALLFLVVIALCLLFFLFT
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PVX_092055 | 420 S | YSRSSYYSN | 0.995 | unsp |