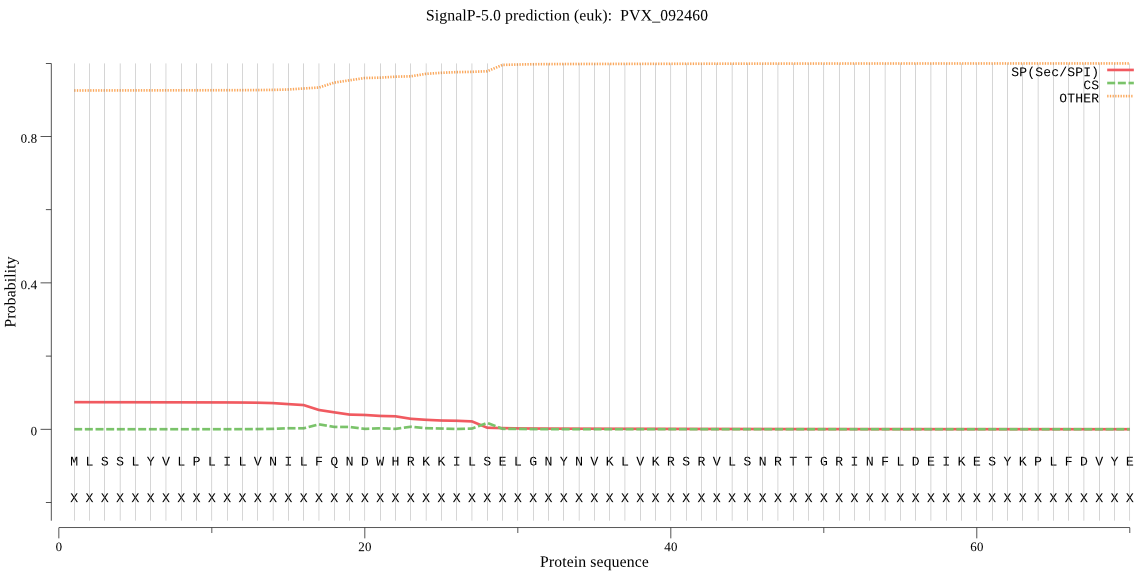
Fasta :-
MLSSLYVLPLILVNILFQNDWHRKKILSELGNYNVKLVKRSRVLSNRTTGRINFLDEIKE
SYKPLFDVYELSAEFEKLRNAKGKGEEKRGRKKKKKKKKKKNDHIGGGDDKETPRLNLIK
LSATQPDGGEHTQKGKSTEAPIREAHSGKSTGINKRAANFIQMLNDGNSNGRGDAEMGVE
NEPDTFSKLINEGNIEDFDHLEKDEPEGEEEKKQEEAGEAEAGAAEGKEKGEEAERKEEQ
EEAKQPQEQEQRDHKSMREEESQSASREEERQSNMRESPPGGESPQEGGDNIEGGTEVAA
RKGGDKSKGQEREGEKNLMASKKEGVKAEKPSEEGSPSGHSLQGEGAEEAPAASPAEDSD
EKKEEEEEEEEEEEEEDSNDQGREYQLNDGNSVAQEDIITQEFDHYTIVTNSNDVLNDIR
VDASDISKLSIESINIPYSEANKTSFTHQRHIVLTNKGNKKYRVVLMTKNPKFIELEDEP
DEKAENNTFIERESMEQKRTPNGRGEHTSDRTNLYGGKGTLDFSKVYSGNGRGSGREVEN
PHVETHTGGGTGEGGIRATINRLFSFLTFGGNPGESARGAGESKDGGGKKESANGGGDRH
HHHYRHDLQSRDNRRSRHDRTKDLNEILSADKLVDQYLLNLKNNTPERQELIFVLRGDLD
LHSTYMRRVIKRSNAKFEEHIKKNFQQVDSIVYDVSSPINFLCFFVPTVFDMSNFHMLKE
ALLVLQKELERYMENWSFSNTYVTLDAPHVGGEGGASAGAGHDGRKDAHMGSRAGDQADG
HADDRETGHTDHREGGAPHGKPRKFVKKKKNLYNVKYSFLRKFWSFDSIIAFARKMNKKN
MDIEKEILNFLPKELREYSTWNLSVIRVFNAWFLAGYGNKNVKVCIIDSGVDKNHIDLMK
NVYIPEYSEQYEMTQDFYDFMVKNPTDSSGHGTHVTGIVGGLANDLGMVGVAPNVTLISL
RFIDGVKYGGSFHAIKAINVCILNKAPIINASWGSSKYDANIYLAVQRLKYTFNGKGTVL
VTAAGNENKNNDEHPLYPSSFKLPHVYSVASISKNFEISPFSNYGVNSVHIMAPGHHIYS
TTPNNSYKINTGTSMAAPHVCGVNALIYSVCYNQGFIPPAEEVLDILTRTSIKVISRRRR
TINDSLVNAEAAVLTTLLGGLWMQMDCHFVKFNLEGGKEKHIPVVFSAYKKGIYETDIVI
AVIPTEENSKVYGEILIPIRIVTDPKAENFKESPRNGKKMIIDENEASHDEVLSYICENA
LYNLYEMDSNFLVMSLVLFFVAFILIVVGTIVFIKRKRHSKYCDDEDHDHNMMRNSVFEH
HHILSGNTNHALKMAKRSHVEKIRNSVRFSVVMGTQNNCVFKERAPKKLDFENYGDLIKK
PLLKSPPKKFVNHRAEQNWGHEAEKKDPRD