

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_092755 | SP | 0.056121 | 0.898278 | 0.045600 | CS pos: 21-22. CRC-FF. Pr: 0.4389 |
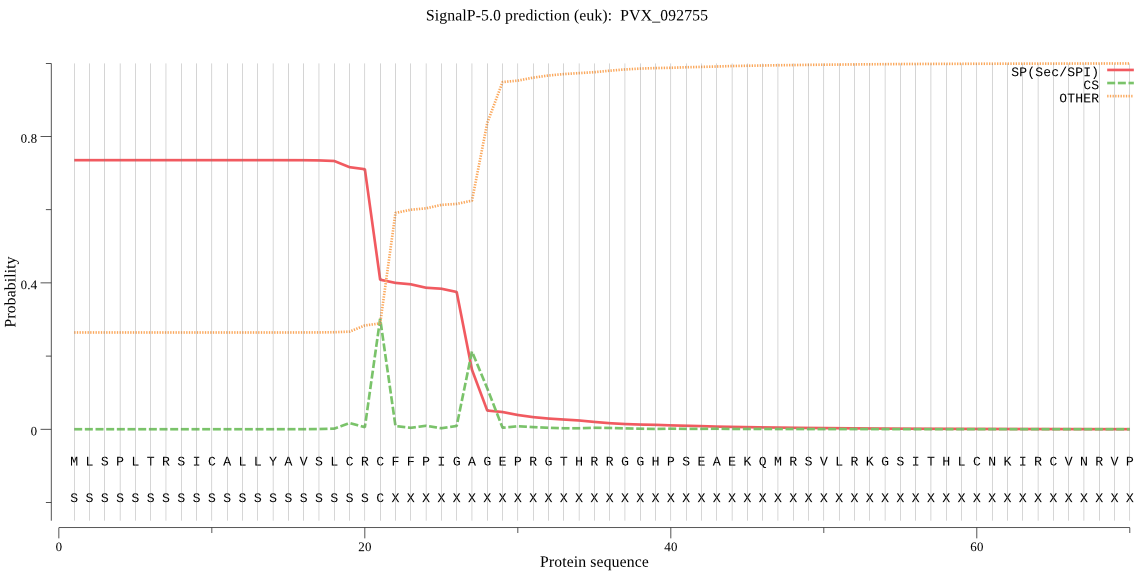
MLSPLTRSICALLYAVSLCRCFFPIGAGEPRGTHRRGGHPSEAEKQMRSVLRKGSITHLC NKIRCVNRVPWNEGKRRISSATLSSGKRSPSSSRLFSSFETRTFGSQAEQVIDGIAFTIY DNTSIYKEETSSTPIVLIHGCYGSKNNFRVFSKSLKSNKIVTIDLRNHGNSKHTDSMKYE EMESDIKKVLNELHIRNCCIVGFSLGGKVSMYCALKNESLFSHLVVMDILPFDYNERKCH VKLPYNISYMTKILFNIKTNLRPRSKAQFLAHLRAQTPGISSSFEQFICTSLREEAVKAV KAGEAAGNAVGADFTGGADAIGCSAGGPSELQNLVWKINVDTIYRELPHILGFPLNHQER KYHNPCSFIIGQKSDLVYTIPQYQSIIKNYFPSSQNFILPEATHTVYIDNAKECADIVNE TLRL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_092755 | 91 S | KRSPSSSRL | 0.996 | unsp | PVX_092755 | 91 S | KRSPSSSRL | 0.996 | unsp | PVX_092755 | 91 S | KRSPSSSRL | 0.996 | unsp | PVX_092755 | 97 S | SRLFSSFET | 0.996 | unsp | PVX_092755 | 154 S | VFSKSLKSN | 0.995 | unsp | PVX_092755 | 176 S | KHTDSMKYE | 0.993 | unsp | PVX_092755 | 85 S | ATLSSGKRS | 0.993 | unsp | PVX_092755 | 89 S | SGKRSPSSS | 0.996 | unsp |