

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
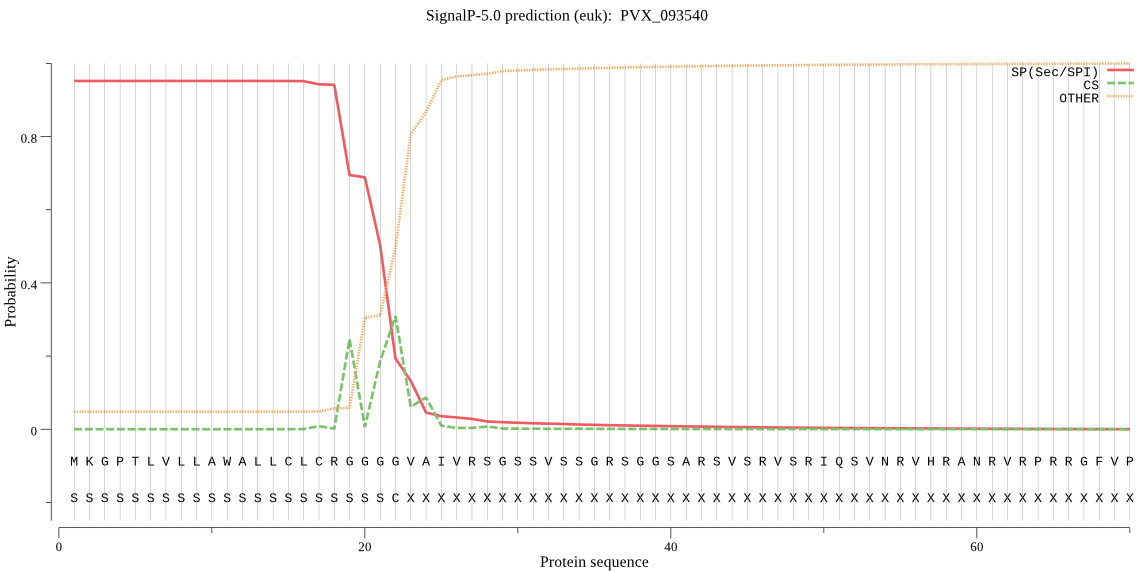
| PVX_093540 | SP | 0.006088 | 0.992624 | 0.001288 | CS pos: 22-23. GGG-VA. Pr: 0.4366 |
MKGPTLVLLAWALLCLCRGGGGVAIVRSGSSVSSGRSGGSARSVSRVSRIQSVNRVHRAN RVRPRRGFVPGGGDSQGSCAKQRRPRRPKLRRNHLHSTSAGEGFSNTVHHDVKKNTKNYS EDGPPASFHRYSENMKTRKNVHPSIRITQFDRTFMKCKEGYNRRYSHLKDSELFGSFRFV GRQKKGITSPRYYLPKYVERPNYHRTGTPVYVPYGSGGQRSGGQMSGSQGSADQRSDFNE RGKSPHPYGNVKREEEIETIRSNCQFARELMDDVSYIICEGVTTNDLDIYILNKCINNGY YPSPLNYHLFPKSSCISINEILCHGIPDNNILYENDIVKVDISVFKDGFHADMCESFLVP KLTKNEKKKKKKFYDFIYLNDKLRTRHTKFILKYHFDLTTNKVVKTGKHCSIKRIRYDPP NSKDRPTEQHRSYDDNSNAVMSSGDLGGNAPFPGGDYHNAADGGALPGEDLDELEQFHRQ YDEQVIYDKQQSSRTYEDLQSYLYRQSRGPPSDHGNRSDHNRFAFFDKRKPGVDELKRLM REKNLDLIRTAHECTMEAIRVCKPGVPFSAIGEAIESHLERKNKGHLRYAVVPHLCGHNI GRNFHEEPFIMHTRNDDDRLLCENLVFTIEPIISERPCDFIMWPDNWTLSNARYFFSAQF EHTIVVRKDGAEVLTGKTDRSPKFVWECEKG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_093540 | 166 S | NRRYSHLKD | 0.993 | unsp | PVX_093540 | 166 S | NRRYSHLKD | 0.993 | unsp | PVX_093540 | 166 S | NRRYSHLKD | 0.993 | unsp | PVX_093540 | 432 S | EQHRSYDDN | 0.994 | unsp | PVX_093540 | 48 S | VSRVSRIQS | 0.996 | unsp | PVX_093540 | 97 S | NHLHSTSAG | 0.993 | unsp |