

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_093555 | OTHER | 0.989178 | 0.003308 | 0.007514 |
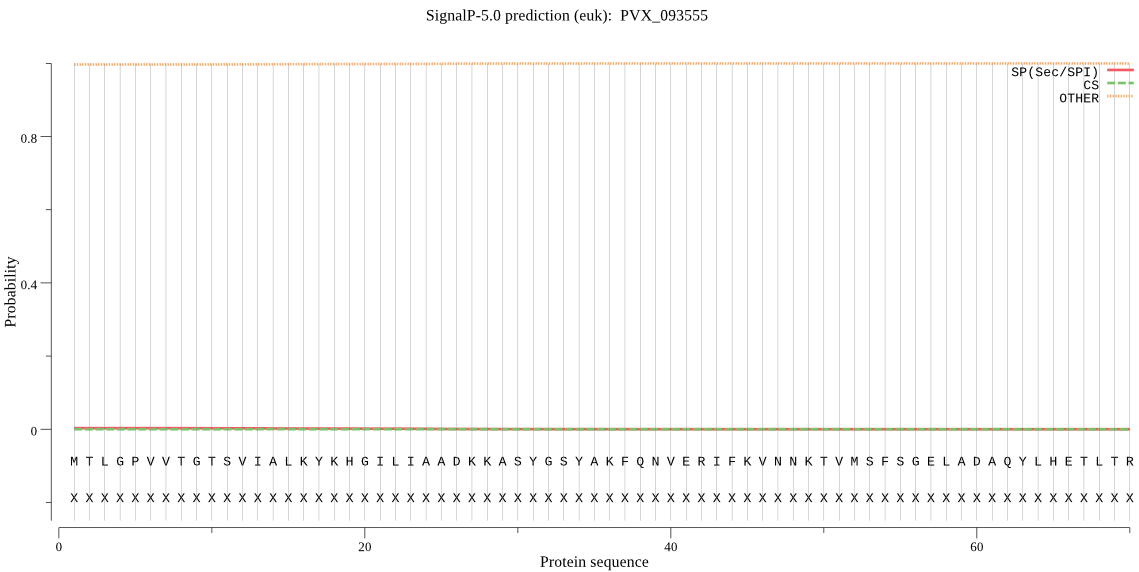
MTLGPVVTGTSVIALKYKHGILIAADKKASYGSYAKFQNVERIFKVNNKTVMSFSGELAD AQYLHETLTRVNVNNVMDKKSKYDLHNSKYYHAYVGRLFYNRKNKIDPLFNTIIVAGLNS QEYDDNDKDVLLYTGKHTTDEYKEINKEDLYIGFVDMHGTQFAGDYMTTGYARYFALTLL RERYKDYMTEEEAKTLLNECLRVLFYRDATASNKIQIVKITSKGVEYEEPYFLSCDVNSR DYVYPSIMLPSTGCMW
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| PVX_093555 | T | 2 | 0.533 | 0.687 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| PVX_093555 | T | 2 | 0.533 | 0.687 |
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PVX_093555 | 81 S | MDKKSKYDL | 0.993 | unsp |