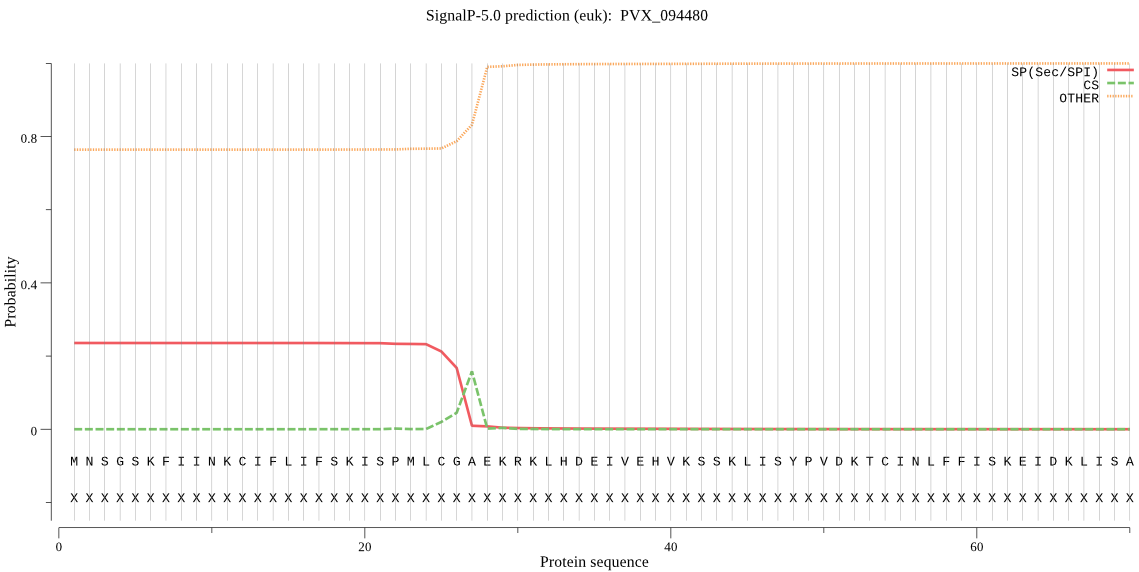
Fasta :-
MNSGSKFIINKCIFLIFSKISPMLCGAEKRKLHDEIVEHVKSSKLISYPVDKTCINLFFI
SKEIDKLISARSAVVSGGDQKRSHTTLQSCLNLYYFLITCAKLKKITFVHKKVLEYVEQR
IATSLQITGVESADEKYIPYSIHIKNEKSVSHLVAELGADCAAYLKKINGVKIRHLTIDR
SQINIDEEWKQKHQCAFNRKKLKIPISKHNYEFILNGVTESALREKVLTLLNSPYENQSL
NRDVIKILKKRYDVATKLGYRNWVDYSMSHFTSEKGKYNKLRDFFNLVKEKVDRDYARVS
VNMGKLLRSGGGSQPSDRYGLPHDRGELRRDRMAPLRDKPTLHDWSYYHNKCANKPDEYF
VNTFFPHGHVWKNFIEIVSRIYNFSYEEVREKGVTKKWPKGTLVYRIERRSANHSSGANT
HVYESLARSRFGESPRREVPPGDSLLGYMYIVPYMEIKLRDYFRVPSMNCLSNTCLICPG
HVVVDYKFIGTIPIEKKLFSCSEILTLFHEFGHAYHLLLLSRRYTLYRFSNMPLDYAEFF
SHMSEHLVNNLSVLQALSNKADNNSKISEDIFKAIKFDSSRLANIFTHAVVDYEIHGLNP
REFFASHLEGRAGVAHGKAHEQVHAQADGQPYDRAHADDSLAPFYNLIGSYLPYKIGPSY
SIHSTSFPYHFSCYYAGSVLSYLLAEMRVLMNIPTADLSIKRNSSLVAFQKKFYAIVAED
FIGSSGWPGSSVIDRALANRHLLA