

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_096240 | OTHER | 0.999979 | 0.000018 | 0.000003 |
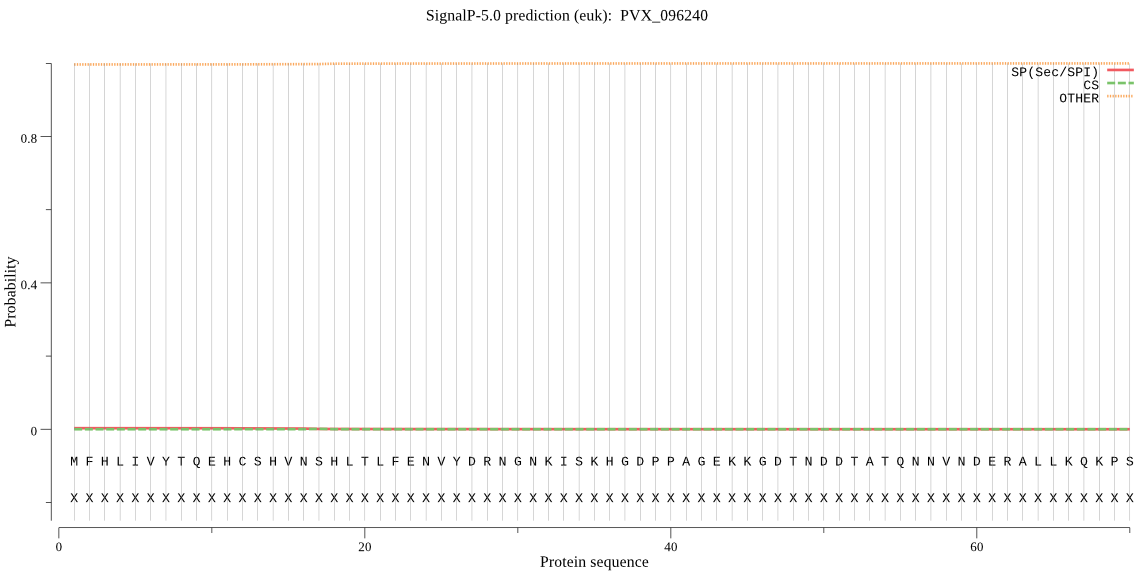
MFHLIVYTQEHCSHVNSHLTLFENVYDRNGNKISKHGDPPAGEKKGDTNDDTATQNNVND ERALLKQKPSQEGGGGPKVQHYTSSFLQLYVNDEKNFDLYLIEEEDNGDGGATSVAASGG GAEDAEAVEVVEAAEAAETAETAETAEAAETAETAEAAETAETAEAAETAEAAEAESEDT RKNQTNSKKGGDKRERGASAKWPKEGTNQKDAQPHEDDKSRRVNLPLHRSHMLFILSLPT HFPFDRLYRLISAYADFIVQVKVFHVRGNICFLDEEESPPGGAAKVESPPGEQSDEGNRR SECNRHSEGNRRSECNRHSEGNRRSECNRRSEDNPHGECPSTPGVEQTAVYQRIHSVVKR RLKKKKKMKSYSVLIFFKTQIYADMFYSEYHCTGLDHLVESHQVEWHNVVDVQGEKGYPD WYVYCAFVSLVYYIVDGEVGTRPVEAEALPQVEESLRRSEGKVSELKKGSKETHGGGNQL PSSSGSNPFDINHFESFQKSVITDGNTCISTCPLCMELLCEEICFTLTRSKKWSERKRGK ASKLRGYLNLSCNVCSLIFLYDSVSRLLGGGALGGAQEGKQTGKQTGVTHSIDNCARDGP GRLGRLVGSLKCRHCSNVDDVWLCLTCANIGCGRYQKSHAKMHSNRYNHHYCLNLKTKKV WNYMREAFIEDKVEDDQSELASAWRGGSGPTGGGFYSSAFHPCVGAAPPGRGDFVEDEYD LCCGDEHPLHPTHDGRICDKIYEIFTDDAYVSDGLKNELLYSLYSLLSHQSNVYNNSLIE LQCSYMNRVDVESRDVQSVRDAVDQVESQNREMEGFLRKAESSIRAKNNERAQLQEKIRF LTELNGNIVSQRRGVSGGDDRDDDSRKIKRLDETIRALQAQVDELLRGLSGGG
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_096240 | T | 154 | 0.541 | 0.059 | PVX_096240 | T | 151 | 0.533 | 0.071 | PVX_096240 | T | 160 | 0.513 | 0.071 | PVX_096240 | T | 145 | 0.509 | 0.059 | PVX_096240 | T | 163 | 0.507 | 0.059 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_096240 | T | 154 | 0.541 | 0.059 | PVX_096240 | T | 151 | 0.533 | 0.071 | PVX_096240 | T | 160 | 0.513 | 0.071 | PVX_096240 | T | 145 | 0.509 | 0.059 | PVX_096240 | T | 163 | 0.507 | 0.059 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_096240 | 199 S | ERGASAKWP | 0.997 | unsp | PVX_096240 | 199 S | ERGASAKWP | 0.997 | unsp | PVX_096240 | 199 S | ERGASAKWP | 0.997 | unsp | PVX_096240 | 455 S | QVEESLRRS | 0.993 | unsp | PVX_096240 | 530 S | TLTRSKKWS | 0.991 | unsp | PVX_096240 | 798 S | RDVQSVRDA | 0.997 | unsp | PVX_096240 | 856 S | RRGVSGGDD | 0.997 | unsp | PVX_096240 | 118 S | SVAASGGGA | 0.993 | unsp | PVX_096240 | 187 S | NQTNSKKGG | 0.995 | unsp |