

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
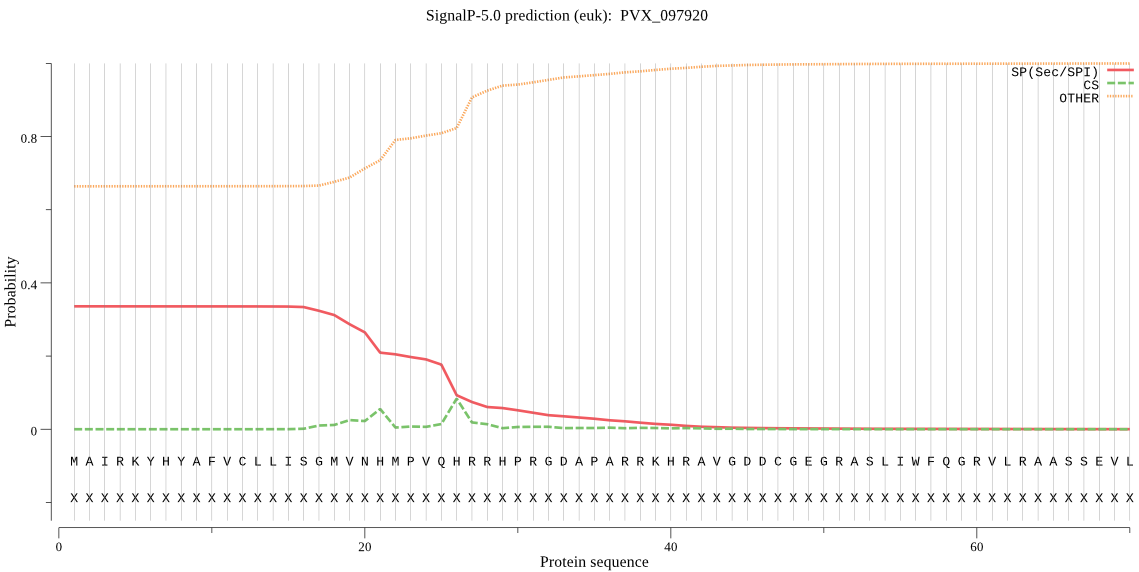
| PVX_097920 | OTHER | 0.751845 | 0.244858 | 0.003297 |
MAIRKYHYAFVCLLISGMVNHMPVQHRRHPRGDAPARRKHRAVGDDCGEGRASLIWFQGR VLRAASSEVLQGGASQPGVSSSSHSSSHSSSHSSSHSSSHSSSHSSSHSSSHSSPHYSPS FWPPPPEEQHHSDEVPTKLRSKLIIKFKQQENAPQFAPMKENLVKLLGSCGRVQKLSHVD LYLYETFPKISERALGKCLQLLSSARVLVEQDQQIYPVEGGCSTGGARGTGGRSGKSVTG GRTPPGSEATFQPNNQLPFKKFLSRLQSDCKGTNIIKGYDQTKMKEGTELSEPHERSDVN VCIVDTGVDYNHQDLQGNVVHVKHGRDGSGDGSGGGGGCDGGGCDFARGMDNHGHGTFIA GIIAGNSQRDSQGIKGICRRAKLTICKALNSKNAGFVSDILNCFNFCASKEAKIINASFA STKNYPSLFEALKTLEQKNILVVSSSGNCCPTAESKNTFTECNLDVMRVYPTAYSTNLRN LITVSNMVQHENGLVTLSPDSCYSSNYVHLAAPGDDIISTFPQNKYAISSGSSFSAAVVT GLAALVLSIKGGLSYEEVIRLLRGSIVQTESLRSKVKWGGFLDVRHLVSAAIALSRASGP AATPSR
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_097920 | S | 95 | 0.632 | 0.023 | PVX_097920 | S | 101 | 0.620 | 0.021 | PVX_097920 | S | 94 | 0.619 | 0.022 | PVX_097920 | S | 105 | 0.619 | 0.021 | PVX_097920 | S | 99 | 0.612 | 0.023 | PVX_097920 | S | 111 | 0.612 | 0.027 | PVX_097920 | S | 98 | 0.610 | 0.022 | PVX_097920 | S | 97 | 0.608 | 0.021 | PVX_097920 | S | 113 | 0.605 | 0.024 | PVX_097920 | S | 110 | 0.603 | 0.022 | PVX_097920 | S | 90 | 0.600 | 0.022 | PVX_097920 | S | 91 | 0.600 | 0.023 | PVX_097920 | S | 93 | 0.599 | 0.021 | PVX_097920 | S | 109 | 0.599 | 0.021 | PVX_097920 | S | 89 | 0.593 | 0.021 | PVX_097920 | S | 103 | 0.591 | 0.023 | PVX_097920 | S | 106 | 0.590 | 0.022 | PVX_097920 | S | 102 | 0.588 | 0.022 | PVX_097920 | S | 114 | 0.586 | 0.028 | PVX_097920 | S | 87 | 0.583 | 0.023 | PVX_097920 | S | 107 | 0.561 | 0.023 | PVX_097920 | S | 118 | 0.553 | 0.078 | PVX_097920 | S | 86 | 0.548 | 0.022 | PVX_097920 | S | 120 | 0.544 | 0.020 | PVX_097920 | T | 239 | 0.532 | 0.079 | PVX_097920 | S | 83 | 0.515 | 0.023 | PVX_097920 | S | 80 | 0.513 | 0.209 | PVX_097920 | S | 85 | 0.512 | 0.021 | PVX_097920 | S | 82 | 0.504 | 0.044 | PVX_097920 | T | 243 | 0.501 | 0.035 | PVX_097920 | T | 230 | 0.500 | 0.088 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_097920 | S | 95 | 0.632 | 0.023 | PVX_097920 | S | 101 | 0.620 | 0.021 | PVX_097920 | S | 94 | 0.619 | 0.022 | PVX_097920 | S | 105 | 0.619 | 0.021 | PVX_097920 | S | 99 | 0.612 | 0.023 | PVX_097920 | S | 111 | 0.612 | 0.027 | PVX_097920 | S | 98 | 0.610 | 0.022 | PVX_097920 | S | 97 | 0.608 | 0.021 | PVX_097920 | S | 113 | 0.605 | 0.024 | PVX_097920 | S | 110 | 0.603 | 0.022 | PVX_097920 | S | 90 | 0.600 | 0.022 | PVX_097920 | S | 91 | 0.600 | 0.023 | PVX_097920 | S | 93 | 0.599 | 0.021 | PVX_097920 | S | 109 | 0.599 | 0.021 | PVX_097920 | S | 89 | 0.593 | 0.021 | PVX_097920 | S | 103 | 0.591 | 0.023 | PVX_097920 | S | 106 | 0.590 | 0.022 | PVX_097920 | S | 102 | 0.588 | 0.022 | PVX_097920 | S | 114 | 0.586 | 0.028 | PVX_097920 | S | 87 | 0.583 | 0.023 | PVX_097920 | S | 107 | 0.561 | 0.023 | PVX_097920 | S | 118 | 0.553 | 0.078 | PVX_097920 | S | 86 | 0.548 | 0.022 | PVX_097920 | S | 120 | 0.544 | 0.020 | PVX_097920 | T | 239 | 0.532 | 0.079 | PVX_097920 | S | 83 | 0.515 | 0.023 | PVX_097920 | S | 80 | 0.513 | 0.209 | PVX_097920 | S | 85 | 0.512 | 0.021 | PVX_097920 | S | 82 | 0.504 | 0.044 | PVX_097920 | T | 243 | 0.501 | 0.035 | PVX_097920 | T | 230 | 0.500 | 0.088 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_097920 | 237 S | RSGKSVTGG | 0.991 | unsp | PVX_097920 | 237 S | RSGKSVTGG | 0.991 | unsp | PVX_097920 | 237 S | RSGKSVTGG | 0.991 | unsp | PVX_097920 | 367 S | IAGNSQRDS | 0.995 | unsp | PVX_097920 | 554 S | KGGLSYEEV | 0.995 | unsp | PVX_097920 | 85 S | SSSHSSSHS | 0.991 | unsp | PVX_097920 | 177 S | VQKLSHVDL | 0.992 | unsp |