

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
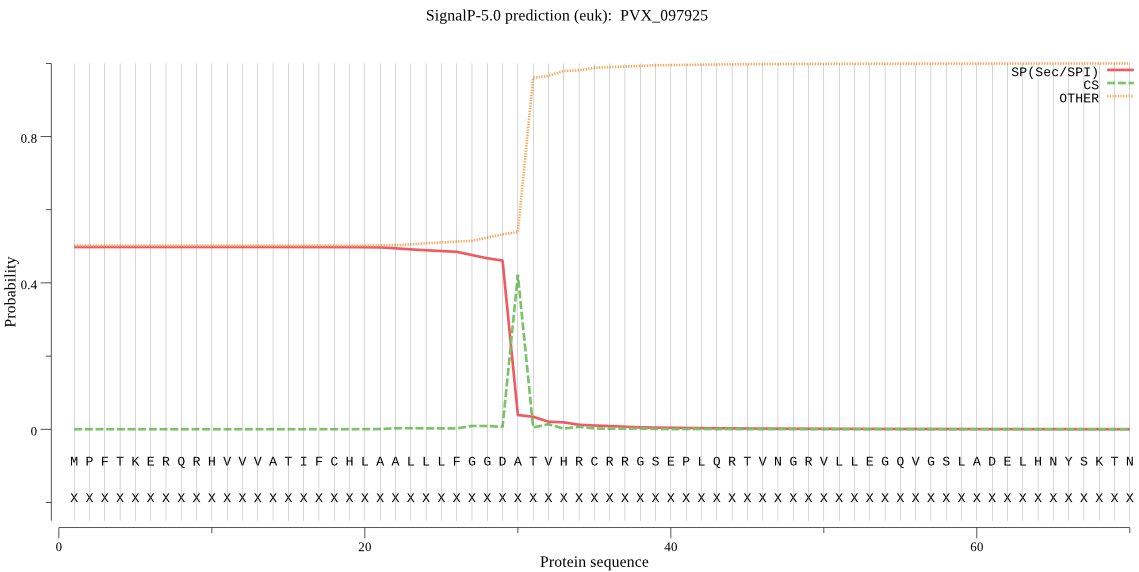
| PVX_097925 | SP | 0.405364 | 0.588948 | 0.005689 | CS pos: 30-31. GDA-TV. Pr: 0.8561 |
MPFTKERQRHVVVATIFCHLAALLLFGGDATVHRCRRGSEPLQRTVNGRVLLEGQVGSLA DELHNYSKTNLSIPSFVQVKLKDSYDRKIRRGGATPEGNYHIPNCDDTDGSANFLKKMYR SIKAFLRITSHEKSWCLKYERFRRVRRDFVYVVQSSLSVGRTRVCLIDTGVDLKDEVLGH FVRMSRGGLNQGGDNPGEQNERRADGINTQRCNEKNYASCQSSDVHDEDMHGTFIANTVI RRDLLMKGGAYKRGVDLIVCNAFAKSFGNAVKNSHLVPLIKCLEMCKERGAKVIHVGYNV QGESEQLVKLMEELQREEIIVVSPSLQVYHRNGGETNSKKEHLEEPSTQKMYPASFADTF ENVFSVGALRNSPQGGLVPILGNANPRGEKPKGEQLHKRENTTLFSFSYGKTFPFGRSPP SMVEDAQAYASADFVNALVMIFNVNPKLSMKRVRLILERSIGRRSELKGLSKWGGYLDPF KLIAETLKERNELCGRFFRELGGNLEEGGGGLLGGGLPRGEATEHLGDWVERPPNDEDQR SCGEATTGEATPRLDAAFPPQGGELEEIERQFEEEAVRGLDHHNLGYYHTDAISKWAEDF PDGGAAPSEGPPSDTTYRSSYNDEEDVYTPDEANQSFYGNSGGVALEVVGEGGLSSLPLG VSFLDKHTSDGGSAVPPLPRRNGRGRGYESGEGTSPPMNRRSLGESGLPERWDQTEGTHV MNEEDRGDGYDGQADWITQMRGTNYVDGHAYKAARVYGNSPEGIPEGELSRPDWGWPTTP LSRWDPRGETPPWGDDHEGYGFSPQLDDDWGGEYGRKGRRRDELGRRLRRDELGRKRTRD ELRRRRRRSPRAVPRQRRRTPTRGRTAKRLTGREAGLVRKNGPTGKSRNGNAMRFRSSGR SVRGRSQRRRPVGGRPVLRKPLLRKPLLRKPILGMPVRAFAPKTSRVVMGRR
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_097925 | T | 615 | 0.513 | 0.019 | PVX_097925 | T | 616 | 0.508 | 0.027 | PVX_097925 | T | 778 | 0.503 | 0.091 | PVX_097925 | T | 779 | 0.500 | 0.028 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_097925 | T | 615 | 0.513 | 0.019 | PVX_097925 | T | 616 | 0.508 | 0.027 | PVX_097925 | T | 778 | 0.503 | 0.091 | PVX_097925 | T | 779 | 0.500 | 0.028 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_097925 | 338 S | GETNSKKEH | 0.993 | unsp | PVX_097925 | 338 S | GETNSKKEH | 0.993 | unsp | PVX_097925 | 338 S | GETNSKKEH | 0.993 | unsp | PVX_097925 | 421 S | RSPPSMVED | 0.994 | unsp | PVX_097925 | 620 S | TYRSSYNDE | 0.997 | unsp | PVX_097925 | 690 S | RGYESGEGT | 0.993 | unsp | PVX_097925 | 702 S | MNRRSLGES | 0.993 | unsp | PVX_097925 | 782 S | TTPLSRWDP | 0.991 | unsp | PVX_097925 | 849 S | RRRRSPRAV | 0.998 | unsp | PVX_097925 | 871 T | AKRLTGREA | 0.99 | unsp | PVX_097925 | 898 S | RFRSSGRSV | 0.994 | unsp | PVX_097925 | 901 S | SSGRSVRGR | 0.992 | unsp | PVX_097925 | 906 S | VRGRSQRRR | 0.996 | unsp | PVX_097925 | 39 S | CRRGSEPLQ | 0.994 | unsp | PVX_097925 | 158 S | QSSLSVGRT | 0.994 | unsp |