

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_097935 | SP | 0.304486 | 0.681112 | 0.014403 | CS pos: 25-26. TLA-GD. Pr: 0.9259 |
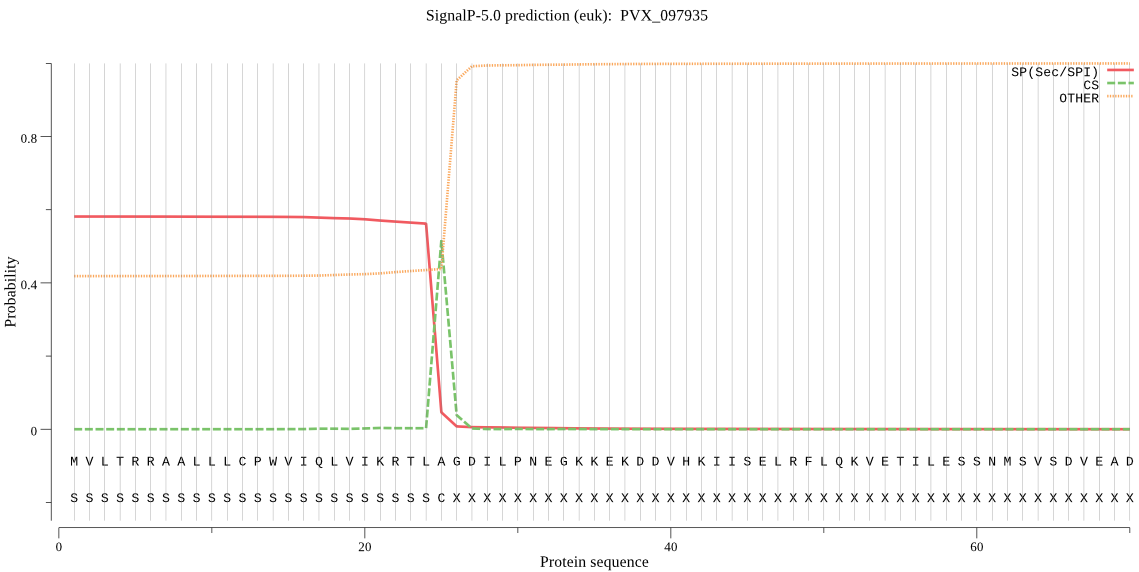
MVLTRRAALLLCPWVIQLVIKRTLAGDILPNEGKKEKDDVHKIISELRFLQKVETILESS NMSVSDVEADANAYNPDRDAPKEELQKLQDQQETPSKEPNNLRNSPQKRAEKKESPGKNK KSLRLIVSENHATSPSFFEESLLQEDVVSFIQSKGKLSNLKNLKSMIIDLNSDMTDEELA EYISLLERKGALIESDKLVGADDVSLASVKDAVRRGESSVNWGKLRSTMLEVPSGESPPS HAASSGSPFDDDDDLLSEAALHREEAHLAGSKTTKGYKFNDEYRNLQWGLDLARLDETQD LINANRVSVTKICVIDSGIDYNHPDLRNNIDVNVKELHGRKGVDDDSNGVVDDVYGANFV NNSGDPMDDNYHGTHVSGIISAVGNNGIGIVGVDGHSKLVICKALDQHKLGRLGDMAHMI NGSFSFDEYSNIFNASVEHLRSLGILFFVSASNCAHDKLSKPDIAKCDLAVNHRYPPILS KTHNNVIAVANLKRDLDESYSLSVNSFYSNIYCQLAAPGTNIYSTTPMNNYRKLNGTSMA SPHVAAIASIVRSINPNLTYLQIVEILRNAIVKLPSLTERVSWGGYVDILRAVNLAIDSK AAPYIKSHSWFRWKQGSRR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_097935 | 115 S | EKKESPGKN | 0.995 | unsp | PVX_097935 | 115 S | EKKESPGKN | 0.995 | unsp | PVX_097935 | 115 S | EKKESPGKN | 0.995 | unsp | PVX_097935 | 134 S | NHATSPSFF | 0.992 | unsp | PVX_097935 | 208 S | VSLASVKDA | 0.998 | unsp | PVX_097935 | 247 S | ASSGSPFDD | 0.997 | unsp | PVX_097935 | 63 S | SSNMSVSDV | 0.996 | unsp | PVX_097935 | 65 S | NMSVSDVEA | 0.991 | unsp |