

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_099107 | SP | 0.391208 | 0.581399 | 0.027394 | CS pos: 18-19. VGC-FN. Pr: 0.6789 |
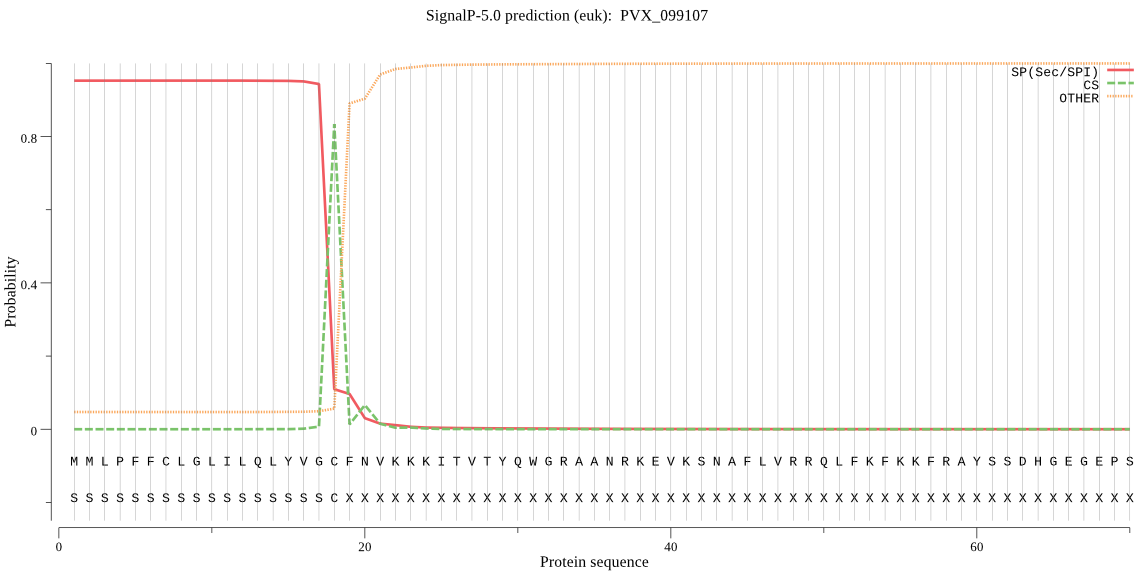
MMLPFFCLGLILQLYVGCFNVKKKITVTYQWGRAANRKEVKSNAFLVRRQLFKFKKFRAY SSDHGEGEPSIDAAEKGANNGATNGATNRAKNTLREFYHRNCSRLRKTLELVKSGLTSIQ DNLMSNSLASQLAISVSFFLLHFYMLSRHFLILFPYQLIPNHSNILMSLDLNNTLFFLMS IFFFKRFKAHFHSFKQRLQLNRFTIKLQENKRKNILSVCFLLIASYVLSGYVSIYTERIL TLCKLFSRPLSDNVVKSLQILTGHFIWVACSIVVFKQLLYPYFLNNESNLNLRHKDSWFF EVIYGYVFSHFVFSVVDLANNFIINRFMGEAEDEMYVDNSIDDIVSGREFVSTLLCIISP CFSAPFFEEFIYRFFVLKSLNLFMHIHYAVTFSSLFFAIHHLNIFNLLPLFFLSFLWSYI YIYTDNILVTMLIHSFWNIYVFLSSLYS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_099107 | 340 S | YVDNSIDDI | 0.991 | unsp | PVX_099107 | 340 S | YVDNSIDDI | 0.991 | unsp | PVX_099107 | 340 S | YVDNSIDDI | 0.991 | unsp | PVX_099107 | 346 S | DDIVSGREF | 0.991 | unsp | PVX_099107 | 93 T | RAKNTLREF | 0.991 | unsp | PVX_099107 | 336 Y | EDEMYVDNS | 0.991 | unsp |