

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
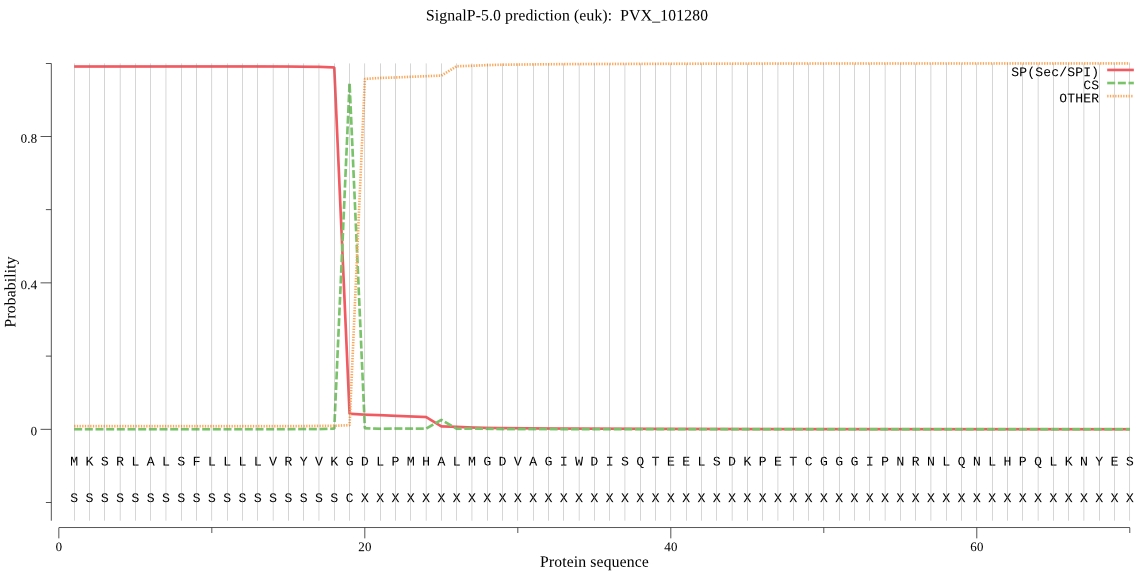
| PVX_101280 | SP | 0.006005 | 0.993598 | 0.000396 | CS pos: 19-20. VKG-DL. Pr: 0.9762 |
MKSRLALSFLLLLVRYVKGDLPMHALMGDVAGIWDISQTEELSDKPETCGGGIPNRNLQN LHPQLKNYESFLEGNYGGLETTSMKLTTEKIKLSERGSQRNNWTYLAVRDVKTNGIVGHW TMVYDEGFEVRVPGRRYFAFFKYERTHSKRCPDPIENKDSTDSKCYKTDPTRTLIGWVLH ERVHPSDKERKAFQWGCFHGKKREEVGVSSFVIHGASGIRGGAQDGEGQFLPPKKQLSFA AVQQRSNYTKIRLSTGGSYGVPKTSLVSIHQRARERIYGCRKEESGEAKIRLTLPKAFTW GDPFSNDHFEEDVEDQMSCGSCYSIATLYSLHKRFEIWLSKKYGKKITMPRLSFQSILSC SPYNQGCDGGFPFLVGKQLYEFGVPTEEAMRYGSSDSIKCEMDMGFYNNVRFAKYKEEDL FFAGEYNYINGCYECSNEFDMMNELYSNGPIVVAINATAKLLSLYNLGKQSGVYDSATHE QQICDVPNEGFNGWQQTNHAVTIVGWGEEHEEGKGGSPVKYWVVRNTWGKAWGYKGYIKF PRGVNLAGIETQAVFLDPDLSRGGAVKMSGHEGAAAGGGAAGGVVGEDGGERGSAMIRPV THTPRAQRGNA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_101280 | 254 S | KIRLSTGGS | 0.996 | unsp | PVX_101280 | 254 S | KIRLSTGGS | 0.996 | unsp | PVX_101280 | 254 S | KIRLSTGGS | 0.996 | unsp | PVX_101280 | 148 S | ERTHSKRCP | 0.997 | unsp | PVX_101280 | 186 S | RVHPSDKER | 0.992 | unsp |