

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
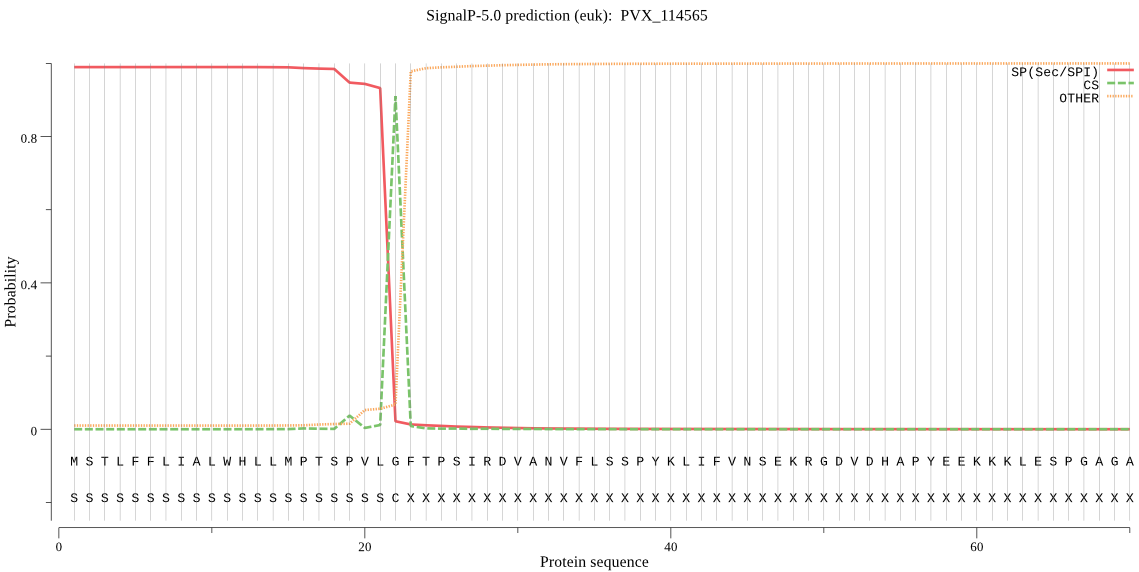
| PVX_114565 | SP | 0.009424 | 0.990171 | 0.000406 | CS pos: 22-23. VLG-FT. Pr: 0.9181 |
MSTLFFLIALWHLLMPTSPVLGFTPSIRDVANVFLSSPYKLIFVNSEKRGDVDHAPYEEK KKLESPGAGAEREEATVSVVQAKPEGEKTQGGEEEKEEKAPKTGVATEEVTAPAGEVTAT QEVVAAAKEAAKVEEVVAATKEAATVEKAATVEKAATVEKAATVEKEATVEKAATVEKAA TVEKAATVEKAATVEKAAATEEAAATEEAAATEEAAATEEAAATEEAAATEEATTTKEAA TTEKAATTEEVAGPAEEAATGEAAAPAGDPARTWKKAKGKSNESETHVVGSPLKKKKYKF FWKQKPNKNKRKGKKKKEPKDESQKKEKKKFPTTILLPGVGGSTLIAEYNNAVIPSCSSN TLNSKPFRLWVSLTRLFSITSNVYCTFDTLRLLYDNEKKIYMNQHGVNITVEDYGRLKGI DYLDYINNTGIGVTKYYHTIAAQFLSKGYVDGESIIGAPYDWRYPLYQQDYNLFKKTIEA AYERRNGMKVNLVGHSLGGLFINYFLVHIVDKKWKQKYLNSILYMSSPFKGTMKTIRALL HGNRDFVSFKITNLIKLSISESMMKAIGNSVGSLYDLIPYKEYYDHDQVVIIMNVDSSPI DENYMQSVVTSCGIYNRDCYLNRTDVKLKVYTLSDWHVLLKGDLMEKYNNHKQYRERHFS MDHGVPIYCVYSALKNKSTDYLLFFQKENLNEEPIIYYGIGDGTVPIESLESCSNFYNAV EKKYFENYSHIGILHSTDAVNYVYDSMQKIEGDLDDTWGDVTTKVAQQDSVQGEAKMKMS PEGSDAGEHDLSTVPQQERGNK
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_114565 | T | 242 | 0.559 | 0.522 | PVX_114565 | T | 241 | 0.555 | 0.070 | PVX_114565 | T | 236 | 0.549 | 0.123 | PVX_114565 | T | 248 | 0.549 | 0.333 | PVX_114565 | T | 247 | 0.541 | 0.062 | PVX_114565 | T | 235 | 0.540 | 0.152 | PVX_114565 | T | 234 | 0.538 | 0.028 | PVX_114565 | T | 260 | 0.537 | 0.254 | PVX_114565 | T | 224 | 0.531 | 0.387 | PVX_114565 | T | 230 | 0.529 | 0.422 | PVX_114565 | T | 212 | 0.520 | 0.387 | PVX_114565 | T | 218 | 0.520 | 0.387 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_114565 | T | 242 | 0.559 | 0.522 | PVX_114565 | T | 241 | 0.555 | 0.070 | PVX_114565 | T | 236 | 0.549 | 0.123 | PVX_114565 | T | 248 | 0.549 | 0.333 | PVX_114565 | T | 247 | 0.541 | 0.062 | PVX_114565 | T | 235 | 0.540 | 0.152 | PVX_114565 | T | 234 | 0.538 | 0.028 | PVX_114565 | T | 260 | 0.537 | 0.254 | PVX_114565 | T | 224 | 0.531 | 0.387 | PVX_114565 | T | 230 | 0.529 | 0.422 | PVX_114565 | T | 212 | 0.520 | 0.387 | PVX_114565 | T | 218 | 0.520 | 0.387 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_114565 | 323 S | PKDESQKKE | 0.994 | unsp | PVX_114565 | 323 S | PKDESQKKE | 0.994 | unsp | PVX_114565 | 323 S | PKDESQKKE | 0.994 | unsp | PVX_114565 | 573 S | NSVGSLYDL | 0.99 | unsp | PVX_114565 | 598 S | NVDSSPIDE | 0.997 | unsp | PVX_114565 | 780 S | KMKMSPEGS | 0.997 | unsp | PVX_114565 | 65 S | KKLESPGAG | 0.994 | unsp | PVX_114565 | 235 T | EEATTTKEA | 0.992 | unsp |