

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_114685 | OTHER | 0.999939 | 0.000027 | 0.000034 |
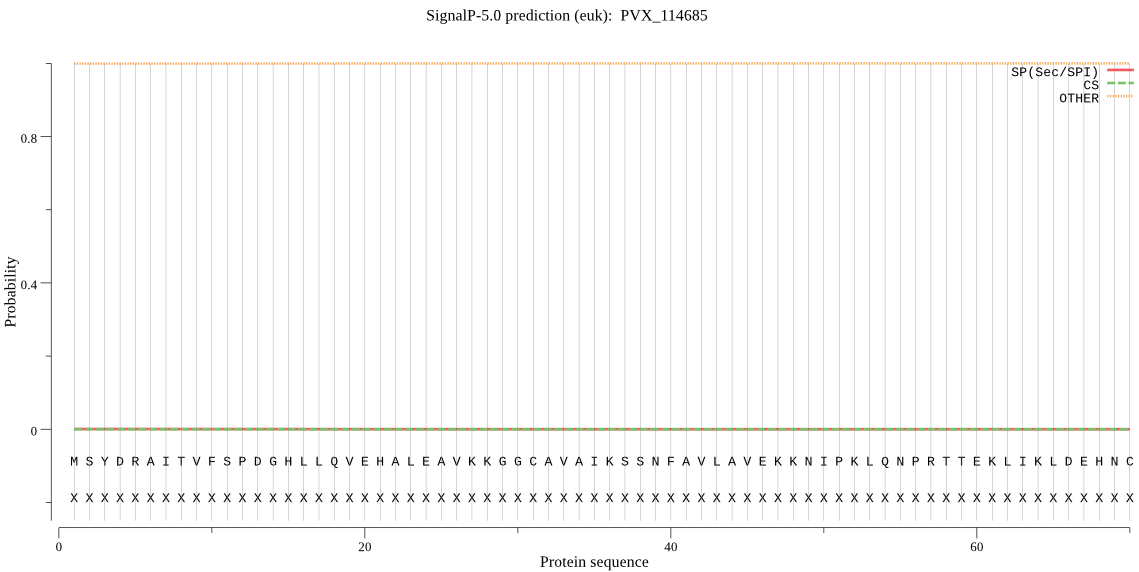
MSYDRAITVFSPDGHLLQVEHALEAVKKGGCAVAIKSSNFAVLAVEKKNIPKLQNPRTTE KLIKLDEHNCLAFAGLNADARVLVNKTRLECQRYFLNMDEPAPVDYIAKYVAKVQQKFTH RGGVRPFGIATLIAGFKNNKEICIYQTEPSGIYASWKAQAIGKNAKVVQEFLEKNYQENM EQNDCLLLAMKAIFEVVELSSKNIEVALLTEKELSFIDEQQINALVEVIDSERTKKNSQN E
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PVX_114685 | 215 S | EKELSFIDE | 0.994 | unsp |