

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_116925 | OTHER | 0.990843 | 0.008869 | 0.000288 |
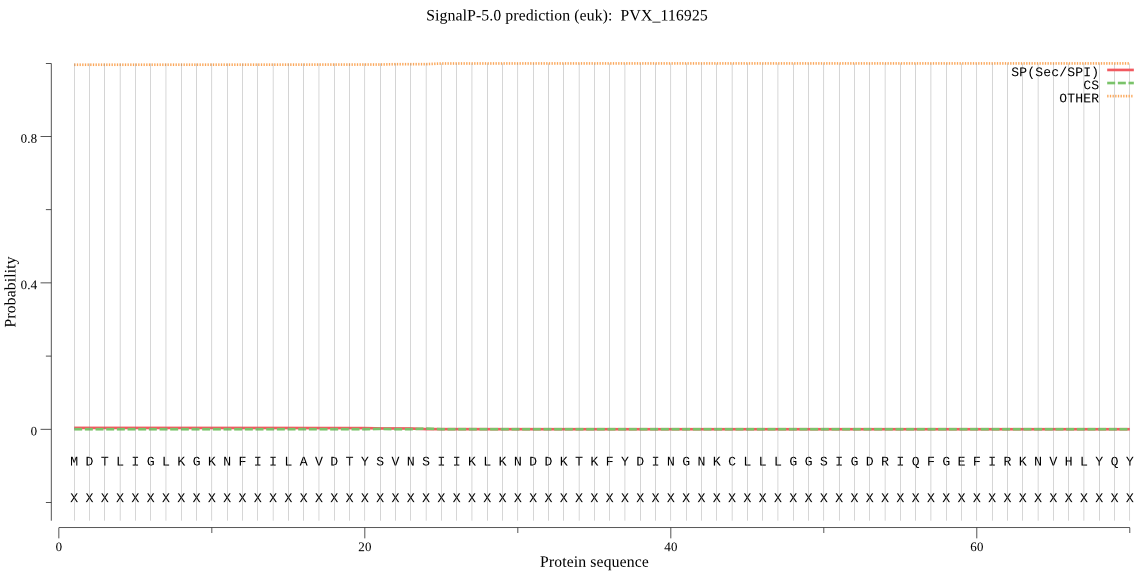
MDTLIGLKGKNFIILAVDTYSVNSIIKLKNDDKTKFYDINGNKCLLLGGSIGDRIQFGEF IRKNVHLYQYQNSTDLFVKSFAFFTRKNLAYYLRRNPYEVNCLIAGYDNKDGYQLYWCDY LSNMDAVNKGAHGYGAYLVNGILDKYYHENMNLEEALLIFKKCFEELKKRFLLTQINYEL RIMADNKIEAQYVTI