

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
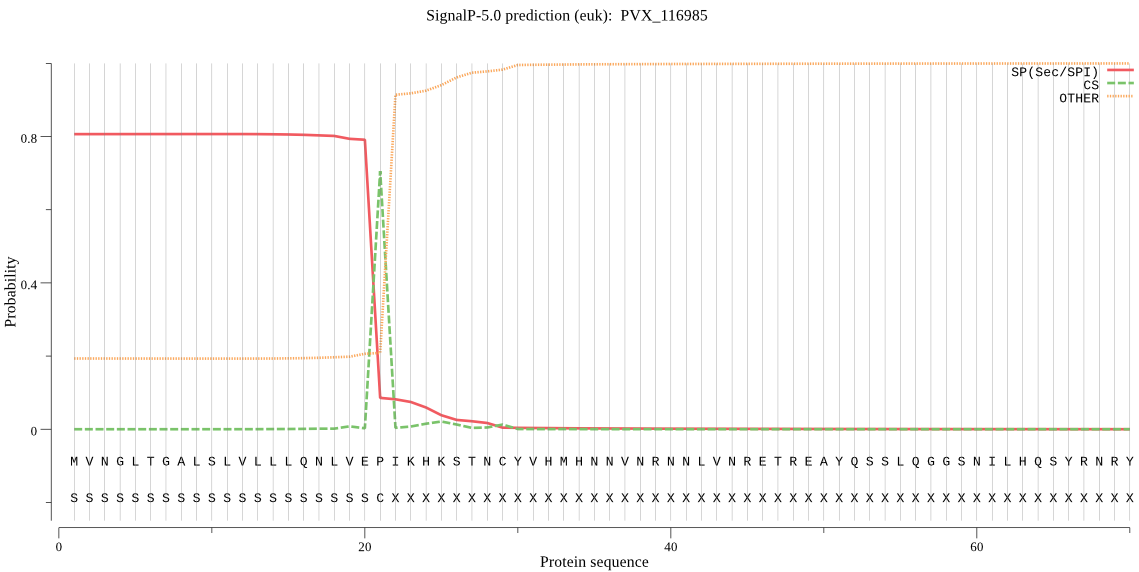
| PVX_116985 | SP | 0.428954 | 0.569312 | 0.001733 | CS pos: 21-22. VEP-IK. Pr: 0.5196 |
MVNGLTGALSLVLLLQNLVEPIKHKSTNCYVHMHNNVNRNNLVNRETREAYQSSLQGGSN ILHQSYRNRYVPLFVSKGTELLNSFFKNNRRGSRHNSTNISKSGKKSAPSSFNESGSSRA TRSGSSHGRKWDPSKCTTREKYSERKSAVEEVADESPNRFSDGQRSSFSDDQGRSPSNHR PSSLSDHTDEGFTSVASNEHHSGNHSPGKSHHPRNMANESGNSDDNPYLNKASYNPGEMP DVFSNWDVAKASVRREKNVQHNFTFMMDLIKESSSMHSKSDDVMIPQNRSGDVLKDGRRL HKGNVKKRKKHLNRLYYSFRGQNDFLSRKGGSHAGDDQSGERFYAYRGDHQAGAHTSDVC HIPVGDSPEQQKEPTVLQRNPHRTKAHSWNSSAIDTAPPASRPNDPADFPTYEEKQRAKE KAHQEYTTYIEERRYPYFNFAKEKNGKIIKKLLIANNGMAAMKCILSIKEWLFKTFSEEN LIKIIVLATEEDISSNAKYISLANKVIKVPPGKNCNNYANVPLIVEVAKKEQVDAVWPGW GHCSENPLLPTMLERENIIFIGPTGDVMEALGDKISANILAQSVNVPVVKWSGDSIRIDN FDSKIGEDVYNKSTIHSLDACIKECQRIGYPVMIKASQGGGGKGIRKVENEAEIKKAYEQ VQMELPNSPIFLMKVCSNVRHIEIQVVGDMYGNVCSLSGRDCTTQRRFQKIFEEGPPSVV PPDIFREMEKASIRLTKMIKYRGAGTIEYLYDQEKQTYFFLELNPRLQVEHPVSEGITDT NLISIQLQVAMGVPLQNIDDIRRLYRIGGGSSGHSVHSVHSGGSGGSVSGGGSVCSGGSV CSGGSERGSQTKECAPPRAEQADAFDFYNHRPHVRNHVIAARITAENSNDSFKPTSGKVQ RIHFQNAKDVWGYFSINDGSVHEFSDSQIGHIFAKGETREVARKNLILSLRKLNIEGEIK TGTRYLAKILECKPFIQNNITTNWLDTIIEKKKNIYYSAVHIIILCATIFKLLIYFTKEK EDIEDALSREDIGIKSQGDSPANLKREVKKAYTFDLVFENVRYNFKAHNTGENLYTLKIN GQEIEVLADYDKKNNKIFASFFNHTYLYTCSEDSLGIHMHLEKDSIFIPKMSNPFHLVSN TNGKIVKYLIKDGEKVGKNEDYIEVEAMKMIMTFKSTEKGILRHRMSEGTIVKVGDLLGV IEQTSHGSGTHPGEQNEIKTFTGHLDLSNKYAYELEEASRLTHPLGFVATDRSTEGIWPI GDQDAGKSHSHMDDANRGEEDTPTVAPNNARSETHRLSEHTGGTHVVEAPKLEDSSSFSS FEQDEVSSRERITKNRLFLKQKKNIFRLFASKRGSDKGEHTAASYLNEKFNSVKAYLTSL SSSSASSAAPTSASTSVKVGEEEVEEAADVEAAVEESAEAAAEAADVEAAQTNKSCANGL SRAHTVNDESGSANRGSSSTGLNSTCQNSLTSERGDRNDHLSSEVVSTRNASNEKASNAG TLSWGAAHRGSSAGASAGASAGESAGVSAGASPGASEGTSKCTSNSCTQVGGDSPGVASP SGHPSGNPNTGEDLRRNQFLLDIPNMKRIEYLLKGYEQDYQQCFNELVKEKKNLNREFAY ILDKFVEYNSLFAQKENITQVEAYDILHTMVKNEKKKNYEIAHAYTYNYLSIKFVELVLK YLLDSGLTPLSEMPQDLLDKLKVISSLRGKCFGNVILLARAIVHIVEHTRLVKLVKCALE DRNATFDEQINQLEELLKMKQRIFAHSKLGKLPMGASSPSSSYGNHCERVVNAFLGSPSD VHMFLPSLFTQENKKALLEIYARNLYTHFDVKAELVGEDCLKFFINNSEEENLLLLNERN RVDVDELISKGGINPTGGLINSIHIINRNGNFLNDYEGGIEHIKKRCKHLYVYHYFGESY GDVYEWVGEGDPGEELHWRQSKNELPHRRTNLLPYQRYILSFERNRQNYDLENVDEKIFK GTKVFLGKYKKNNKIESLFAERVIHLSELPESGASELRETPPKGGNTLGEQNRLEDALAE EFQEAVKDVAIARLNPKVQDAKVSSYITYHVVLEEVKGKDELDVIVEALKNFLNRHNEML LENYVNDVYACVYRKGGESAKGRPQLAPQKVFRLHVLFDGESNVIEEVEGVPPFDVDALY LKRKRAREIDTLYAYDFVKVINISLNRMNGGGPGDLFTYVNSVREFRLDLEALQGGNTTK EGHTTKEGHTTKEGHTTKEGHTTKEGHTTKEGHTTKEGHTTKEGHTTKEGHTTKEGHTTK EGHTTKEGHTTKEGHTTKEGHTTKESHTTKEGHTTKESHTTKESHTTKESHTTKEDATRA DLLLPRQDGLSEDEAKIRKALHLSDELNVGQNKLSVIGLLMNVKTDEYKEGRDVIFIIND ITTHGGSFSVVEDQLFYAISSYAREKKIPRIYISCNSGARIGLYNFLMDKIQVSWKDEEK KELGYDYIYITEEVKSRIAKEDIIFLKEITQNGEKRYIVEGIVGNLNSHIGVENLRGSGL IAGETSKAYDEIFTLSYVTGRSVGIGAYLVRLGKRTIQKKGSSLLLTGFNALNKILGEKV YVSNEQLGGVKIMMRNGISQLEAQNDQDGVDKIFKWLSYVPKTSDHYYDVIEKALEGRSE IINYERANGEEAHNMVREPSQVESQPTPQKEPPKSYFQRINDIDMDGLQNADIVELISGT DSKQGFLDKHSYFEYMNEWGKGILTGRGKLGSIPVGVIAVNRNLVTQVTPCDPALKTKAV RSTHAPCVFFPDNSYKTAQSIEDFNKENLPLFVFANWRGFSGGTMDMFNSVLKFGSMIVN QLVNYKHPVFVYIPIFGELRGGSWVVVDETLNSQIIEMYADTNSKGGILEPPGIVEVKFR LPEIRKLMHGIDSSIIALDERLAEAQEPDDVSRIKQEIEEKEKELLPFYLQVCHRYADLH DVSKCMKSKGVIRKIVPWEKARSFFYYRLVRRLLMSTLSKKYGTALTQSEEFKKLTSDIA SSDEEDYEVCRRILNESTLRDIERLTQDLQHRKTLDEFYRAFQSLPPQQRRELFSKLSSH E
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PVX_116985 | 117 S | NESGSSRAT | 0.992 | unsp | PVX_116985 | 117 S | NESGSSRAT | 0.992 | unsp | PVX_116985 | 117 S | NESGSSRAT | 0.992 | unsp | PVX_116985 | 143 S | REKYSERKS | 0.992 | unsp | PVX_116985 | 147 S | SERKSAVEE | 0.997 | unsp | PVX_116985 | 167 S | GQRSSFSDD | 0.995 | unsp | PVX_116985 | 175 S | DQGRSPSNH | 0.994 | unsp | PVX_116985 | 183 S | HRPSSLSDH | 0.997 | unsp | PVX_116985 | 252 S | VAKASVRRE | 0.992 | unsp | PVX_116985 | 278 S | SSMHSKSDD | 0.997 | unsp | PVX_116985 | 367 S | PVGDSPEQQ | 0.997 | unsp | PVX_116985 | 388 S | TKAHSWNSS | 0.994 | unsp | PVX_116985 | 849 S | SERGSQTKE | 0.993 | unsp | PVX_116985 | 891 S | NSNDSFKPT | 0.994 | unsp | PVX_116985 | 896 S | FKPTSGKVQ | 0.995 | unsp | PVX_116985 | 1028 S | EDALSREDI | 0.998 | unsp | PVX_116985 | 1036 S | IGIKSQGDS | 0.994 | unsp | PVX_116985 | 1270 S | GKSHSHMDD | 0.995 | unsp | PVX_116985 | 1319 S | SSSFSSFEQ | 0.995 | unsp | PVX_116985 | 1355 S | SKRGSDKGE | 0.997 | unsp | PVX_116985 | 1396 S | SASTSVKVG | 0.997 | unsp | PVX_116985 | 1487 S | SEVVSTRNA | 0.991 | unsp | PVX_116985 | 1492 S | TRNASNEKA | 0.993 | unsp | PVX_116985 | 2119 S | KGGESAKGR | 0.992 | unsp | PVX_116985 | 2351 S | QDGLSEDEA | 0.997 | unsp | PVX_116985 | 2454 S | KIQVSWKDE | 0.998 | unsp | PVX_116985 | 2660 S | VREPSQVES | 0.994 | unsp | PVX_116985 | 2698 S | VELISGTDS | 0.995 | unsp | PVX_116985 | 2780 S | KTAQSIEDF | 0.991 | unsp | PVX_116985 | 3001 S | SDIASSDEE | 0.993 | unsp | PVX_116985 | 3002 S | DIASSDEED | 0.997 | unsp | PVX_116985 | 93 S | NRRGSRHNS | 0.996 | unsp | PVX_116985 | 103 S | NISKSGKKS | 0.996 | unsp |