

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PVX_117040 | OTHER | 0.999789 | 0.000185 | 0.000027 |
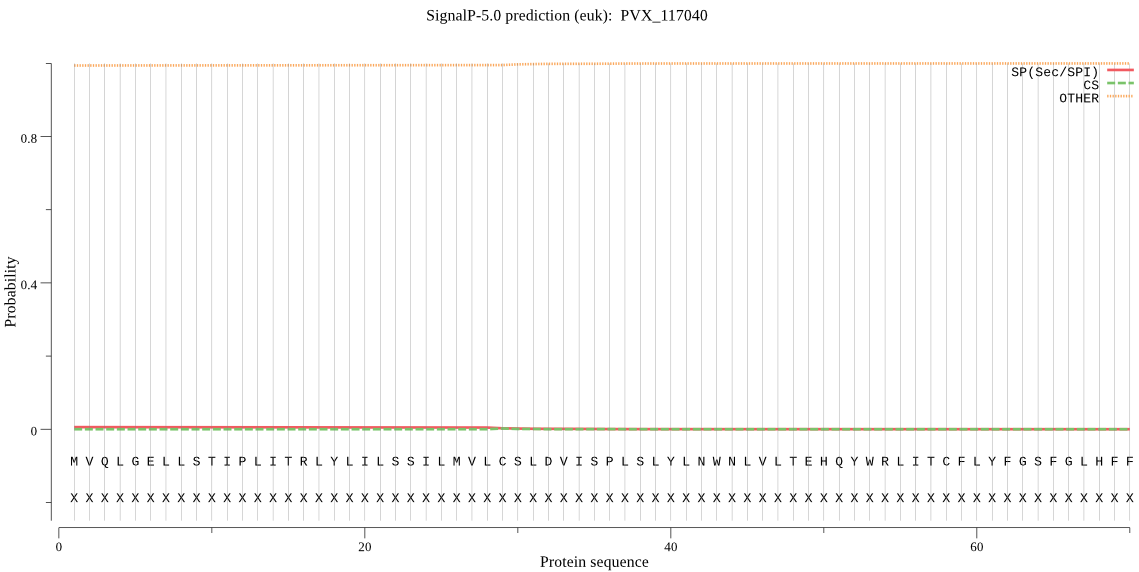
MVQLGELLSTIPLITRLYLILSSILMVLCSLDVISPLSLYLNWNLVLTEHQYWRLITCFL YFGSFGLHFFWDAYVLIYYCSSLEDVTFRNNSADFLWMIIVSCMMLLAVSYLFGGVYFYS SCIINVITYVWSKNNSSTRLTIFFFTIKASYLPWVLTLLSLIVDYNSNDNFFGILVGHIY FFFTNVFPLMPVAKNTQIFKTPQILKWLLKQEQ